| i | Metric | Scale | Organism | Correct (%) | Misclassified mito | Misclassified ER | Miclassified total |
|---|---|---|---|---|---|---|---|
| 1 | Patch 11 | TM Tendency | H. sapiens | 82 | 26 | 12 | 38 |
| 2 | TMD | Kyte & Doolittle | H. sapiens | 82 | 33 | 5 | 38 |
| 3 | Segment 19 | Kyte & Doolittle | H. sapiens | 81 | 29 | 9 | 38 |
| 4 | Segment 11 | TM Tendency | H. sapiens | 81 | 27 | 12 | 39 |
| 5 | Segment 15 | TM Tendency | H. sapiens | 81 | 25 | 14 | 39 |
| 6 | Wheel face 9 | TM Tendency | H. sapiens | 81 | 26 | 13 | 39 |
| 7 | Wheel face 7 | TM Tendency | H. sapiens | 81 | 27 | 13 | 40 |
| 8 | Segment 11 | Kyte & Doolittle | H. sapiens | 81 | 28 | 12 | 40 |
| 9 | Segment 15 | Kyte & Doolittle | H. sapiens | 81 | 30 | 10 | 40 |
| 10 | Patch 19 | TM Tendency | H. sapiens | 80 | 27 | 14 | 41 |
| 11 | Patch 15 | TM Tendency | H. sapiens | 80 | 30 | 11 | 41 |
| 12 | Segment 19 | TM Tendency | H. sapiens | 80 | 26 | 15 | 41 |
| 13 | Patch 19 | Kyte & Doolittle | H. sapiens | 80 | 30 | 12 | 42 |
| 14 | Patch 11 | Kyte & Doolittle | H. sapiens | 79 | 28 | 15 | 43 |
| 15 | Patch 15 | Kyte & Doolittle | H. sapiens | 78 | 28 | 17 | 45 |
| 16 | TMD | TM Tendency | H. sapiens | 78 | 31 | 14 | 45 |
| 17 | Wheel face 5 | TM Tendency | H. sapiens | 77 | 28 | 19 | 47 |
| 18 | Patch 15 | TM Tendency | S. cerevisiae | 88 | 7 | 2 | 9 |
| 19 | Wheel face 5 | TM Tendency | S. cerevisiae | 87 | 6 | 4 | 10 |
| 20 | Wheel face 7 | TM Tendency | S. cerevisiae | 87 | 6 | 4 | 10 |
| 21 | Patch 11 | TM Tendency | S. cerevisiae | 87 | 6 | 4 | 10 |
| 22 | TMD | TM Tendency | S. cerevisiae | 87 | 6 | 4 | 10 |
| 23 | Segment 15 | TM Tendency | S. cerevisiae | 86 | 6 | 5 | 11 |
| 24 | Segment 19 | TM Tendency | S. cerevisiae | 86 | 6 | 5 | 11 |
| 25 | Patch 19 | TM Tendency | S. cerevisiae | 84 | 7 | 5 | 12 |
| 26 | Segment 11 | TM Tendency | S. cerevisiae | 83 | 6 | 7 | 13 |
4 Sequence-based features that are determinant for tail-anchored membrane protein sorting in eukaryotes
Adapted from Fry MY*, Saladi SM*, Cunha A, Clemons WM. 2021b. Sequence-based features that are determinant for tail-anchored membrane protein sorting in eukaryotes. Traffic 22:306–318. doi:10.1111/tra.12809
4.1 Abstract
The correct targeting and insertion of tail-anchored (TA) integral membrane proteins is critical for cellular homeostasis. TA proteins are defined by a hydrophobic transmembrane domain (TMD) at their C-terminus and are targeted to either the ER or mitochondria. Derived from experimental measurements of a few TA proteins, there has been little examination of the TMD features that determine localization. As a result, the localization of many TA proteins are misclassified by the simple heuristic of overall hydrophobicity. Because ER-directed TMDs favor arrangement of hydrophobic residues to one side, we sought to explore the role of geometric hydrophobic properties. By curating TA proteins with experimentally determined localizations and assessing hypotheses for recognition, we bioinformatically and experimentally verify that a hydrophobic face is the most accurate singular metric for separating ER and mitochondria-destined yeast TA proteins. A metric focusing on an 11 residue segment of the TMD performs well when classifying human TA proteins. The most inclusive predictor uses both hydrophobicity and C-terminal charge in tandem. This work provides context for previous observations and opens the door for more detailed mechanistic experiments to determine the molecular factors driving this recognition.
Keywords: co-chaperones, EMC, GET pathway, protein targeting, SND pathway, tail-anchored proteins
4.2 Introduction
Biogenesis of membrane proteins is an essential yet complicated process necessary for maintaining cellular homeostasis. Synthesized by ribosomes in the cytosol, membrane proteins account for approximately a third of the proteome and must be targeted to specified membranes (reviewed in (Fry and Clemons, 2018; Guna and Hegde, 2018; Krogh et al., 2001)). A hydrophobic alpha-helical stretch, often a transmembrane domain (TMD), encodes this information and its position within an open reading frame dictates the cellular machinery responsible for its recognition and targeting (Guna and Hegde, 2018). While computational methods have refined the ability to detect and predict cellular localization of these integral membrane proteins over time (Almagro Armenteros et al., 2017), the precise molecular signals continue to be elusive. Historically, decoding known signals into detailed rules has proven difficult given their great variation and the lack of sequence motifs–thus these signals are often discussed at a high level, for example, hydrophobic alpha-helical stretches. Despite the inability to define these rules, cellular chaperones accurately recognize the various signals to sort substrates into their distinct cellular destinations.
Here, we attempt to address one class of membrane proteins, tail-anchored (TA) proteins, found across cellular compartments and involved in a variety of roles including vesicle trafficking, protein translocation, quality control and apoptosis (reviewed in References (Borgese et al., 2003; Chartron et al., 2012; Fry and Clemons, 2018; Rabu et al., 2009)). TA proteins are marked by a single TMD near their C-terminus and account for approximately 2% of the genome (Chartron et al., 2012; Denic, 2012; Kutay et al., 1993; Wattenberg and Lithgow, 2001). Because of the position of their signals, TA proteins are translated by the ribosome and then post-translationally targeted primarily to the endoplasmic reticulum (ER) or outer mitochondrial membrane. The TMD and C-terminal residues following have been demonstrated to be necessary and sufficient for correct targeting in many experimental contexts (Lin et al., 2021; Wang et al., 2010). Thus, it is suggested that the information recognized by TA protein targeting pathways is contained within the TMD and neighboring residues.
The recent identification of a new route for TA proteins to the ER membrane has challenged how we previously differentiated between mitochondria and ER-bound TA proteins (Guna and Hegde, 2018; Rao et al., 2016; Wattenberg and Lithgow, 2001). To date, while the cellular components involved in mitochondrial TA protein targeting remain unclear, multiple overlapping pathways have been identified for TA protein targeting to the ER membrane (Aviram et al., 2016; Chartron et al., 2012; Fry and Clemons, 2018; Guna et al., 2018; Schuldiner et al., 2008; Stefanovic and Hegde, 2007). The first identified and most studied pathway is the Guided Entry of TA protein (GET) pathway (Schuldiner et al., 2008; Stefanovic and Hegde, 2007). Consisting of six proteins, Sgt2 and Get1–5, the GET pathway is responsible for targeting ER-bound TA proteins (“ER TA proteins” for simplicity) with more hydrophobic TMDs. In yeast, the co-chaperone Sgt2 first captures TA proteins from Ssa1 and, with the aid of Get4 and Get5, transfers the client to the ATPase Get3 that acts as the central targeting factor of the pathway (Chio et al., 2017; Cho and Shan, 2018; Guna and Hegde, 2018; Shao and Hegde, 2011a, 2011b). An ER membrane bound Get1/2 complex facilitates disassociation of the Get3/TA complex and insertion of the TA protein into the membrane. Recently, Guna and colleagues demonstrated that human Get3 (HsGet3) fails to bind to TA proteins with relatively low hydrophobicity within their TMDs. These proteins instead are inserted into the ER membrane by the ER Membrane Complex (EMC) (Guna et al., 2018). A 10-subunit complex, the EMC inserts TA proteins delivered by calmodulin. For TA proteins with moderately hydrophobic TMDs, both the GET pathway and EMC can facilitate insertion. A third dedicated pathway capable of targeting TA proteins into the ER membrane is the SRP-independent (SND) pathway (Aviram et al., 2016). Snd1, the first component of the SND pathway, interacts with the ribosome and possibly the nascent chain while the membrane bound Snd2 and Snd3 interact with the translocon complex. In the absence of the GET pathway, the SND pathway is capable of targeting ER TA proteins with TMDs further away from their C-termini. These overlapping pathways, dependent on either hydrophobicity or signal positions, highlight the diversity in these proteins and the difficulty in identifying a common characteristic of ER-destined TMDs (Aviram et al., 2016).
General patterns have been observed based on exploration of targeting information within the TMD and the C-terminal residues of TA proteins. ER TA proteins tend to have more hydrophobic TMDs (Chitwood et al., 2018; Guna et al., 2018; Rao et al., 2016; Wattenberg and Lithgow, 2001) while some mitochondria TA proteins are amphipathic (Wattenberg and Lithgow, 2001). By modifying the positive charge following their TMDs with an example TMD, studies have shown how insertion by the GET pathway into the ER membrane can be impaired (Figueiredo Costa et al., 2018; Rao et al., 2016). Distinction between peroxisomal and mitochondria TA proteins have been made based on the charge of their C-terminal tails, whereas mitochondria and ER TA proteins in mammals are differentiated by a combination of TMD hydrophobicity and C-terminal charge (Costello et al., 2017). A charged tail was overcome by increasing the hydrophobicity of the TMD, directing the mitochondrial TA protein to the ER. Guna and colleagues determine a threshold in total hydrophobicity by modifying a model TMD to delineate substrates that are inserted either via the GET or EMC pathways (Guna et al., 2018). Throughout these previous works, the ability of these rules to separate ER vs mitochondrial TA proteins at-large has not been systematically assessed, so their broader applicability is still unclear.
With multiple pathways with overlapping substrates, understanding the factors within substrates recognized for targeting is critical. Here we show that formalizing previously suggested criteria, while adequate, are not sufficient for classifying ER TA proteins with moderately hydrophobic TMDs suggested to be substrates of the EMC insertase. We demonstrate through computational and experimental methods that classifying TA proteins by the presence of a hydrophobic face in their TMD is more inclusive, properly capturing both ER TMDs with low hydrophobicity and mitochondrial TA proteins in both yeast and humans.
4.3 Results
4.3.1 Curating TA proteins with experimentally determined localizations
In order to screen TA proteins to identify a concise criterium for localization, we first curated a comprehensive set of TA proteins from the yeast proteome pulling together localizations across public repositories and publication-associated datasets. We screened the reference yeast genome from UniProt (UniProt Consortium, 2021) for putative TA proteins and filtered for unique genes longer than 50 residues (Figure 4.1A). Uniprot and TOPCONS2 (Tsirigos et al., 2015) were used to identify proteins with a single TMD within 30 amino acids of the C-terminus (Borgese et al., 2003) that lacked a predicted signal peptide (as determined by SignalP 4.1 (Nielsen, 2017)). While this set encompasses proteins previously predicted as TA proteins (Beilharz et al., 2003; Kalbfleisch et al., 2007), it is larger (95 vs 55 or 56) and we believe a more accurate representation of the repertoire of TA proteins (Figure 4.1A). Based on their UniProt-annotated and Gene Ontology Cellular Components (GO CC) localizations (Ashburner et al., 2000; Gene Ontology Consortium, 2021), TA proteins were subcategorized as ER-bound (encompassing labels including cell membrane, Golgi apparatus, nucleus, lysosome and vacuole membrane and referred to as ER TA proteins), mitochondrial (inner and outer mitochondria membrane [IMM & OMM]), peroxisomal, and unknown (Figure 4.1C). This set is readily available for future analyses (Table 4.2). The majority of proteins have no annotated cellular localization. Several previously identified TA proteins are not identified by our pipeline and excluded from this new set. These proteins include OTOA (otoancorin) that contains a predicted signal peptide, FDFT1 (squalene synthase or SQS) with two predicted hydrophobic helices by this method, and YDL012C which has a TMD with very low hydrophobicity (Beilharz et al., 2003; Guna et al., 2018). This analysis was also applied to the human genome and a list of 573 putative TA proteins was compiled and annotated based on published localizations (Figure 4.1A-C). Like with the yeast list, the human list is larger than previous reports (573 vs 411), and the majority of the proteins have no annotated localization.
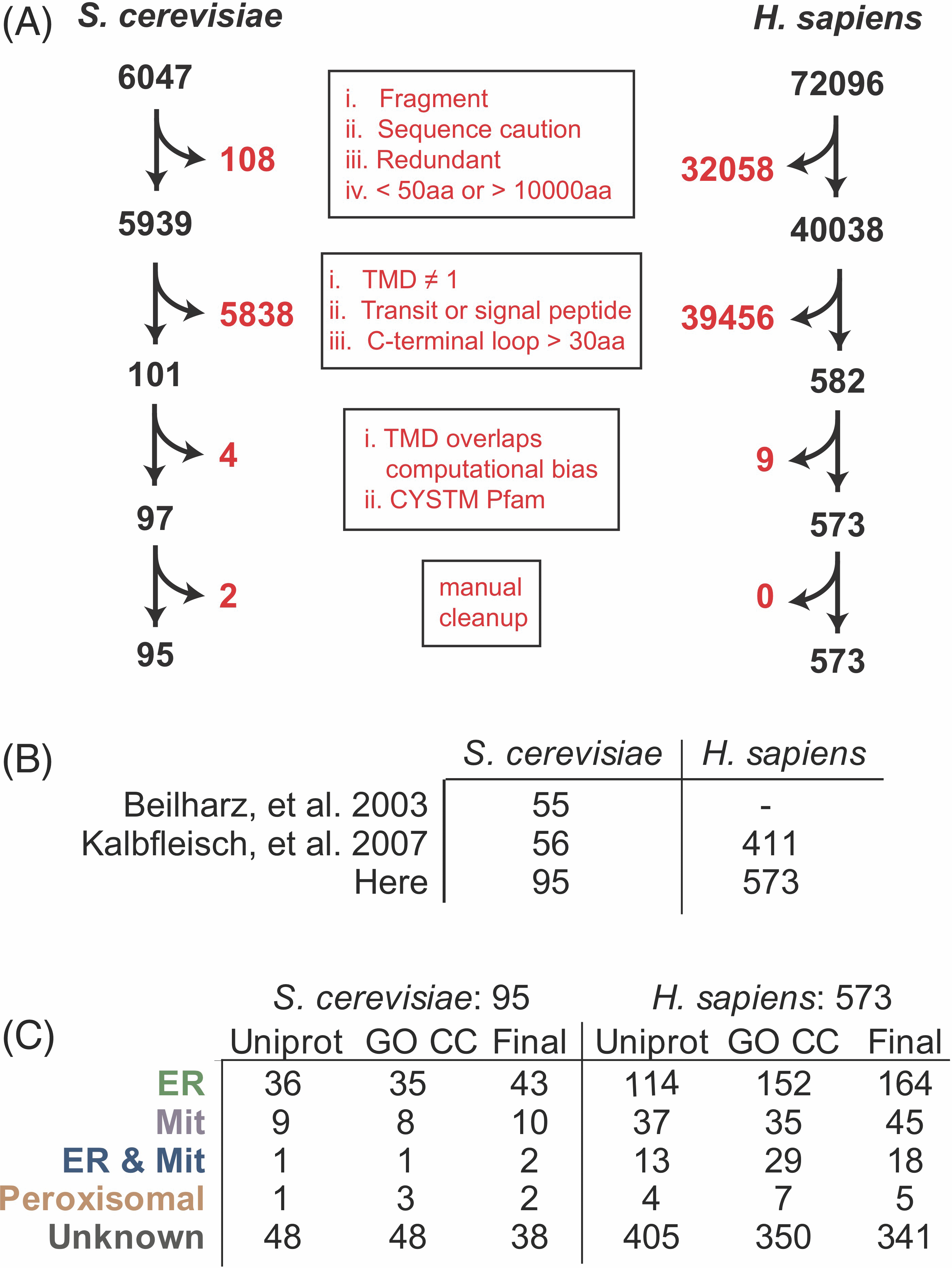
4.3.2 Assessing current metrics for TA classification
To identify factors encoded within TA proteins that ensure correct localization, we began by considering several posited properties including the charge following the TMD, TMD length and TMD hydrophobicity. Previous reports suggest that the presence of positively charged residues following the TMD of mitochondria-bound TA proteins prevents insertion into the ER membrane (Figueiredo Costa et al., 2018; Rao et al., 2016). The number of positively charged C-terminal residues for all 95 yeast proteins was calculated, avoiding issues associated with defining the extent of TMDs by counting any charge from the center of the predicted TMD to the C-terminus. No clear separation is observed when plotting TA proteins with known localizations by number of positively charged residues (Figure 4.2A). As a metric this does a poor job distinguishing between the two; six ER-annotated proteins have a C-terminal positive charge of three or more and one out of the eight mitochondria-annotated proteins has no C-terminal positive charge. Furthermore, neither negative nor net charge of the C-terminal loop separates ER from mitochondrial TA proteins (Figure 4.2B,C). While modulating the C-terminal positive charge affects localization (Rao et al., 2016), cells do not solely use this signal to specify protein localization. Considering the difference in lipid compositions of the ER and mitochondrial membranes, a signal might be encoded in the TMD lengths, but this metric also fails to separate the two sets (Figure 4.2D).
TMD hydrophobicity is the proposed localization-determining feature of TA proteins in studies thus far (Guna and Hegde, 2018; Rao et al., 2016; Wattenberg and Lithgow, 2001). The TM tendency scale, used here and in past studies with TA targeting (Guna et al., 2018; Guna and Hegde, 2018), is a statistical hydrophobicity scale that incorporates both hydrophobicity and helical propensity into a single value assigned to each of the 23 amino acids by using amino acid propensities in TMDs known at the time of its creation (Zhao and London, 2006) (Figure 4.2E). The total hydrophobicity (sum of each residue’s hydrophobicity value) of a TMD sufficiently splits ER and mitochondrial proteins but places a significant number of ER TA proteins among mitochondrial TA proteins. In other words, the total hydrophobicity can classify GET pathway substrates as ER-bound but fails to identify substrates of the EMC insertase that are also ER TA proteins (Guna et al., 2018). For example, the TMD of squalene synthase, a bona fide EMC substrate (Guna et al., 2018), has a lower hydrophobicity than that of model mitochondrial TA protein, Fis1 (Total TM Tendency = 12.5 vs 18.78, respectively). Limiting the hydrophobicity to a single helix stretch, that is, 18aa, sees no improvement in classification (Figure 4.2F).
To examine this inability to correctly classify lower hydrophobic ER TA proteins, we comprehensively assess hydrophobicity across a variety of established scales (Eisenberg et al., 1984; Fauchere and Pliska, 1983; Roseman, 1988; Wimley et al., 1996; Zhao and London, 2006) (Figure 4.2G) and then quantitatively assess predictive power using the receiver operating characteristic (ROC) framework (for a primer, see Reference (Swets et al., 2000)). An ROC curve captures how well a numerical score separates two categories, here ER vs mitochondria, and whose figure of merit is the area under the curve (AUROC). This is a more accurate representation of prediction than simpler numbers like accuracy and precision, which require setting a specific threshold in a numerical score because it accounts for sensitivity and selectivity. A perfect separation gives an AUROC of 100 whereas a random separation results in an AUROC of 50. No matter the hydrophobicity scale used, the total hydrophobicity captures the ER vs mitochondria split to varying extents. In each case, the mean hydrophobicity performs more poorly, yet considering the most hydrophobic 18-residue single-helix stretch results in a slight improvement in predictive ability suggesting that a subset of the helix can explain recognition (Figure 4.2G).
4.3.3 TMD residue organization better classifies TA protein localization
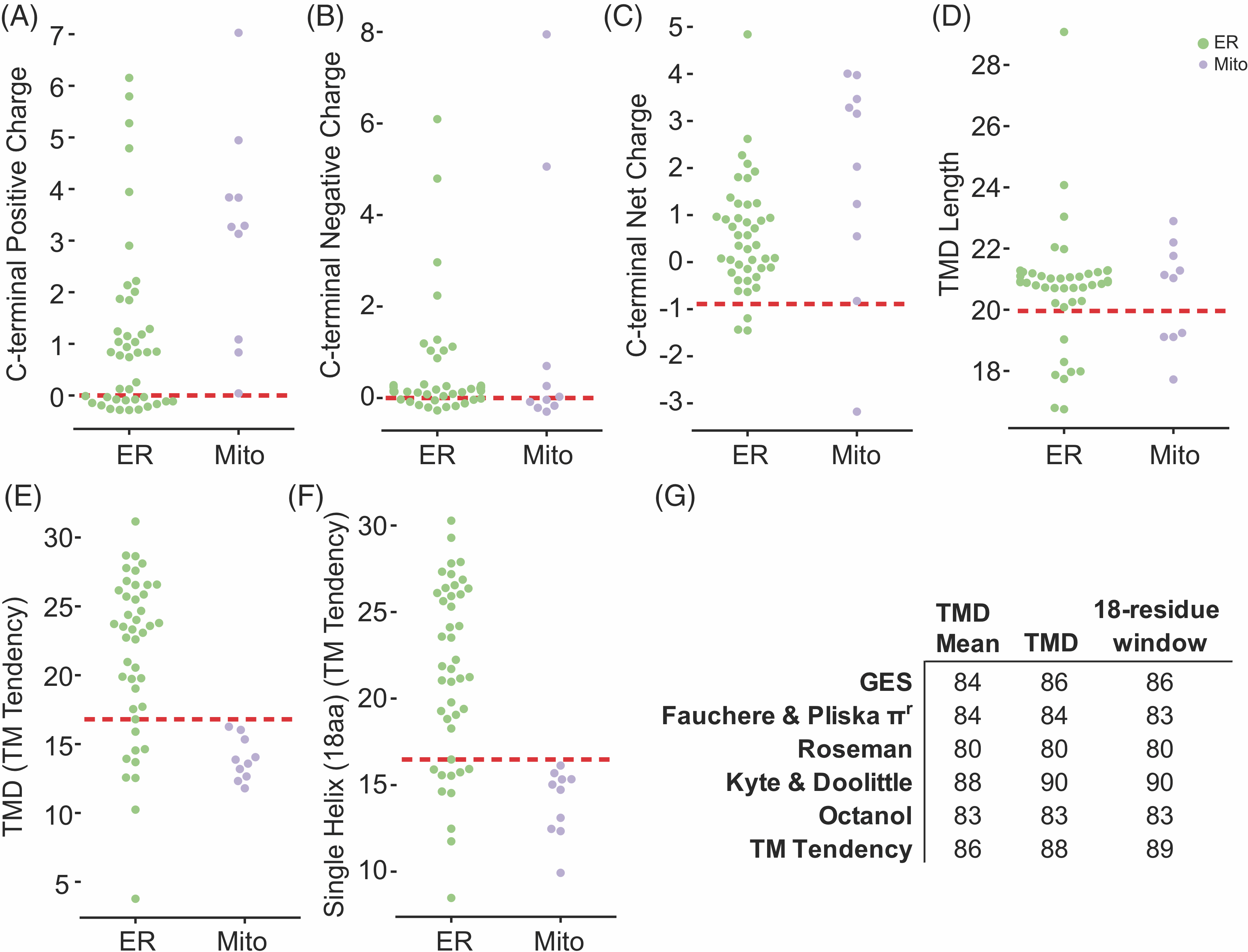
We wondered if TA protein classification could be improved by carefully assessing the hydrophobicity of the TMDs. Data showing that Sgt2 (a co-chaperone in the GET pathway) binds a TMD of a minimal length of 11 residues suggests only a subset of each helix may be necessary to classify localization (Lin et al., 2021). Indeed, the maximum hydrophobicity of segments, specified by the number residues selected, better classifies ER vs mitochondrial TA proteins across hydrophobicity scales (Figure 4.3A,B).
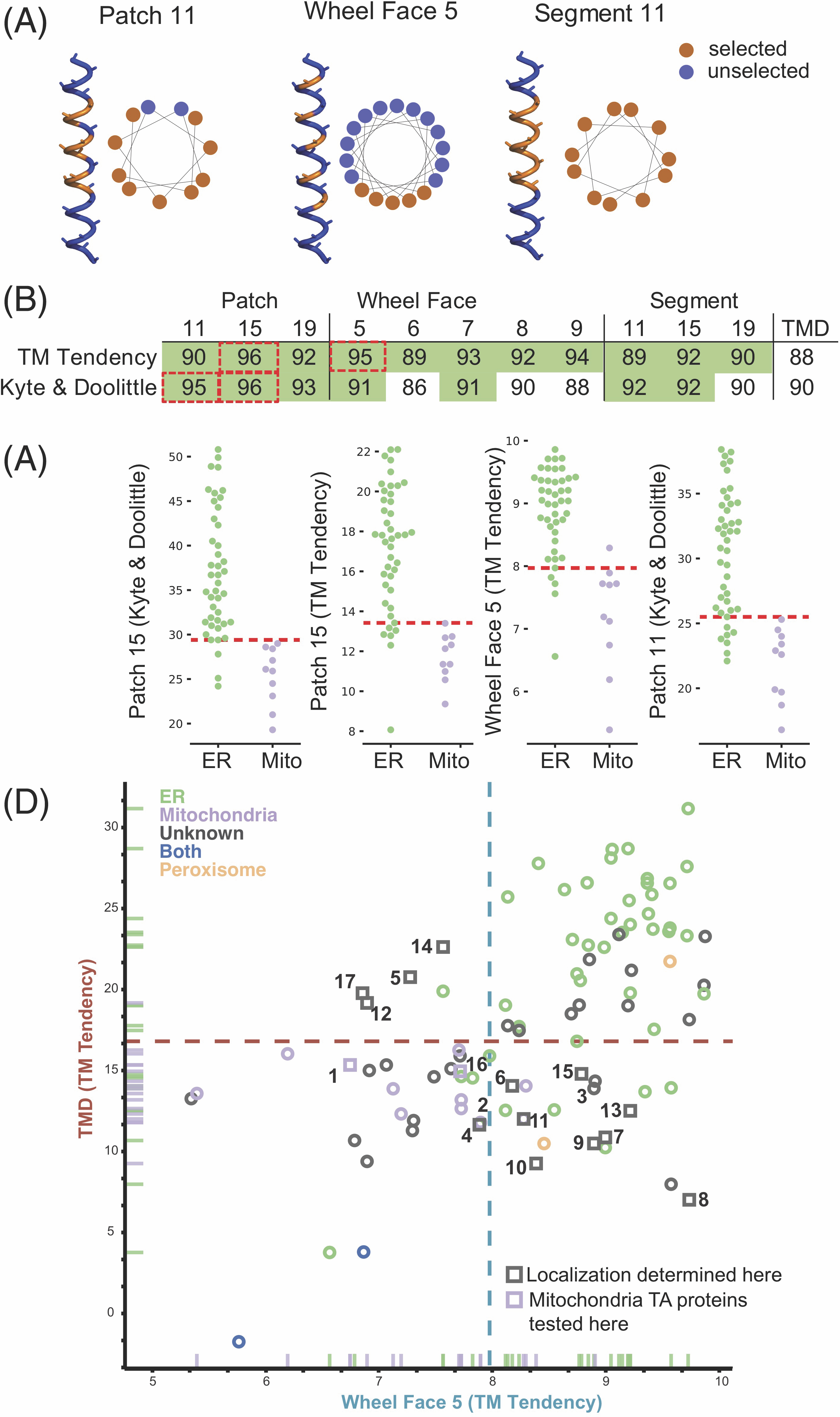
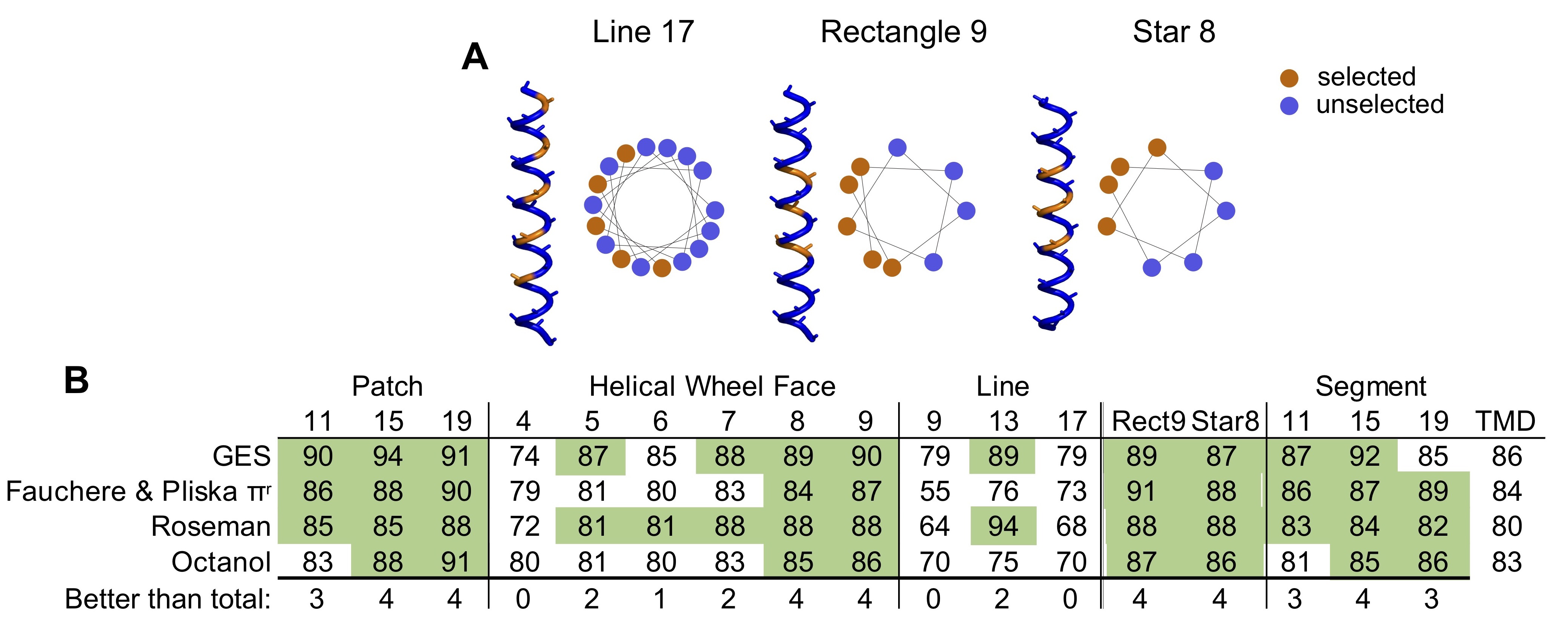
Furthermore, it was also reported that TMDs where the most hydrophobic residues cluster to one side of a helical wheel plot (Schiffer and Edmundson, 1967), a 2D representation of an alpha-helix, bind more efficiently to Sgt2 (Lin et al., 2021). We sought to examine if this clustering is a feature of ER TA proteins and absent in mitochondria TA proteins. This clustering we define as a helical wheel face (Wheel Face) and specify a length by the number of residues selected (Figure 4.3A,B). We also extend the face along the sides of the helix, defining a Patch, selecting three of the four residues in a single turn of a helix. Patch geometries are specified by length of the segment considered, that is, Patch 11 is confined in a 11 segment residues with 9 residues selected (Figure 4.3A,B). Improvements in classification over the total hydrophobicity metric are seen in several cases (Figure 4.3B, green, Supplementary Figure 4.1B, green, Table 4.3). The metrics with the best classification capability are Patch 15 (Kyte & Doolittle and TM Tendency), Wheel Face 5 (TM Tendency scale) and Patch 11 (Kyte & Doolittle scale; Figure 4.3B, dashed red box). These metrics have an improved AUROC value of 96, 96, 95 and 95, respectively, compared to the TMD hydrophobicity score of 90 (Kyte & Doolittle) and 88 (TM Tendency; Figure 4.3B). At the best threshold of the ROC curve, these metrics correspond to five, seven, six and eight miscategorized proteins, respectively. A scatter plot illustrates how these metrics translate to improved separation of ER and mitochondrial TA proteins (Figure 4.3C).
Other hydrophobic geometries were also explored as potential competing hypotheses: residues in a line (every fourth residue), rectangle (one residue plus two residues two away on either side) or star (two adjacent residues and one residue two away on either side; Supplementary Figure 4.1A,B). As with the Patch geometries, these geometries are specified by the length of the TMD considered. Again, improvements are seen in geometries that present hydrophobic patches, that is, Rectangle 9 and Star 8, where line geometries rarely improved classification regardless of scale used (Supplementary Figure 4.1B). Given the relative dearth of experimental data and the substantial number of hypotheses being tested (geometries and hydrophobicity scales), it is difficult to definitively say if one geometry is the sole deciding factor for localization based only on bioinformatics. Regardless of the hydrophobicity scale used, it is clear that the organization of hydrophobic residues within a TMD is important for targeting TA proteins to their intended membranes.
4.3.4 Testing the localization of unknown TA proteins
We then tested if either face (Wheel Face or Patch), Segment, or TMD hydrophobicity metrics enabled us to predict the localization of unknown TA proteins. To do this we selected a subset of unknown TA proteins, whose localization would be predicted differently by TMD and Wheel Face 5 metrics using the TM Tendency scale (Figure 4.3D, numbered gray points, Table 4.4). This selection was made because of the strong AUROC and biochemical data suggesting TA protein containing a helical wheel face bind more efficiently to Sgt2. Several in this group have a hydrophobicity less than the previously suggested cut-off for EMC substrates (Guna et al., 2018) (Figure 4.3D, lower right quadrant). Our experimental setup based on that from Rao et al–GFP is fused N-terminally to the TMD and C-terminal residues of the unknown TA protein (Figure 4.4A yellow panel). Localization is determined by overlap with either a BFP-tagged mitochondria presequence that marks the mitochondria (Figure 4.4A cyan panel) and a tdTomato-tagged Sec63 acting as an ER marker (Figure 4.4A magenta panel).(Rao et al., 2016) Overlap was determined computationally using two algorithms we developed: one to segment individual cells in brightfield and another to determine which fluorescence probe the GFP overlapped with on a per cell basis (Table 4.4).
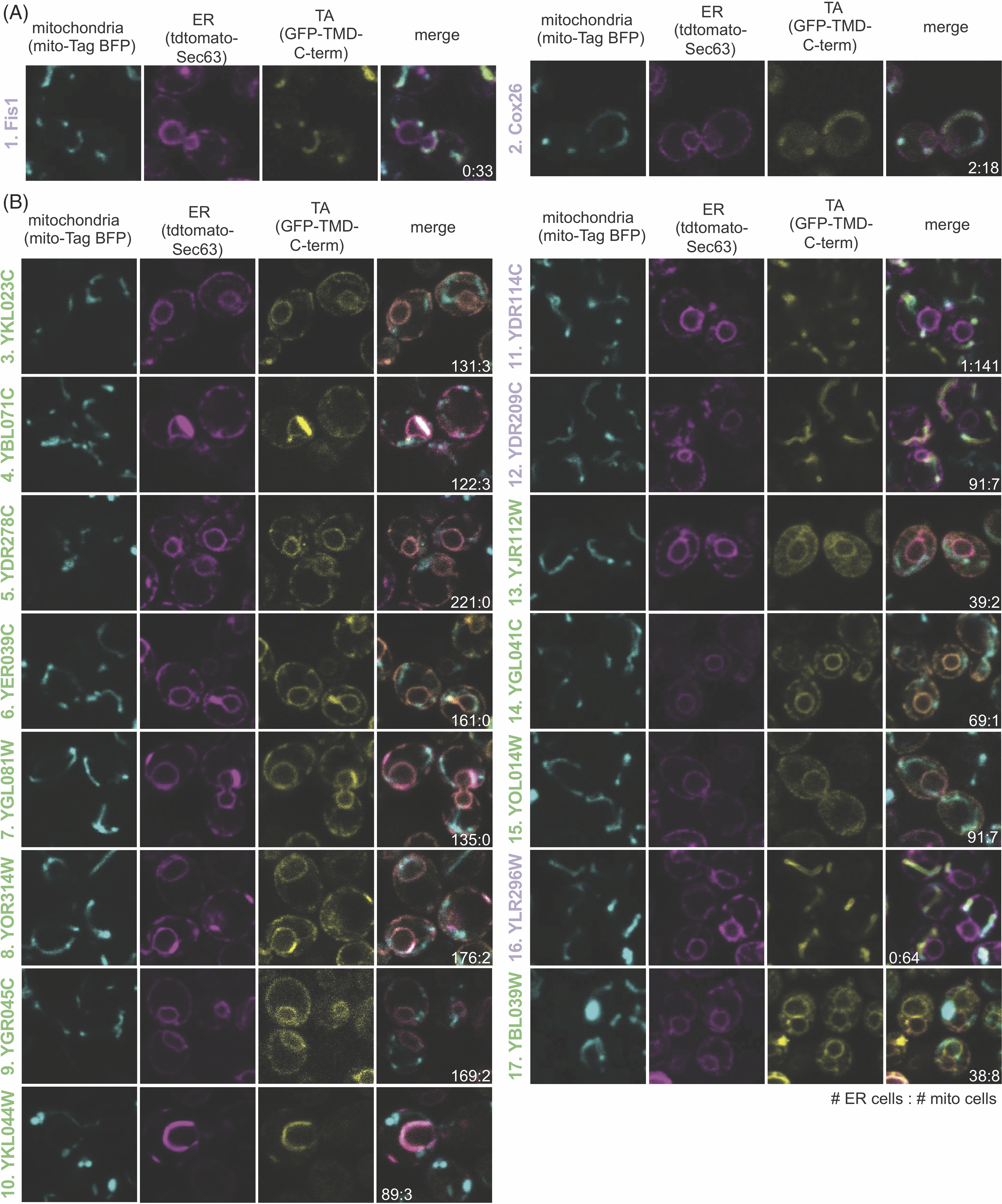
This experimental setup and computational analysis were first applied to the known mitochondria proteins Fis1 and Cox26 (Hartley et al., 2019; Levchenko et al., 2016; Rao et al., 2016). The analysis correctly determines these proteins to colocalize with BFP, thus correctly classifying them as mitochondria TA proteins (Figure 4.4A). We then experimentally tested the 15 Unknown TA proteins where 11 localize to the ER, three to the mitochondria, and one to another cellular compartment (Figure 4.4B, Table 4.1). The localization of this latter TA protein cannot be determined by our experimental setup except to say it does not clearly colocalize with the ER or mitochondria markers visually or through our computational analysis (Table 4.4). The shape of the organelle is consistent with localization to the ER-derived vacuole (Figure 4.4B, #17) (Vida and Emr, 1995). In total, we report the first localization of 10 previously Unknown TA proteins.
Several datasets report protein localizations in yeast but are not yet, or partially, integrated into bioinformatics databases like Uniprot. One in particular was of use for this study, reporting the localizations assigned by qualitatively accessing the pattern of protein expression in images of 17 TA proteins in the Unknown category (Weill et al., 2018) (Table 4.5). Coincidentally, a few of these proteins were included in our experimental test set, for a combined 27 new TA proteins with previously unknown localizations (Table 4.4 and Table 4.5). Of the TA proteins identified by Weill et al. (2018), all but one, YKL044W, was confirmed (Table 4.4). Given the ability to mark ER and mitochondria and quantitate colocalization on a per-cell basis, we use the localization determined here throughout our analysis, that is, YKL044W localizes to the ER. Collectively, we have compiled a list of 27 TA proteins and their localizations that have yet to be integrated into protein databases or reported: 20 ER, six mitochondrial, and one peroxisomal.
4.3.5 Reassessing classification metrics using newly determined localizations
The newly determined localizations were compared to the predicted localizations of the best performing hydrophobicity metrics. Total hydrophobicity metrics across all scales only correctly predict 9 or 14 of the 26 ER and mitochondria TA proteins. Experimental localizations from this work and the Schuldiner Lab (Weill et al., 2018) result in a putative yeast TA protein list with 88% having known localizations (Figure 4.5A). With most localizations known, comparing metrics based on AUROC values is a good representation of the overall dataset (Table 4.6). The best performing metrics were Wheel Face 7 (TM Tendency) and Wheel Face 5 (TM Tendency), with scores of 89 and 88, respectively vs the TMD hydrophobicity AUROC score of 76 (Table 4.6). These metrics correctly predicted the localization of 19 out of 26 and 17 out of 26, respectively, of the subset of our test set that localized to the ER or mitochondria (Figure 4.5A,B). A Patch geometry using the Fauchere & Pliska scale performs well when predicting new localizations–correctly predicting 18 of 26 localizations (Figure 4.5A). Segment metrics performed similarly when predicting new localizations and their AUROC values improved with the inclusion of the new localizations (Figure 4.5A). In all, metrics focused on the organization of hydrophobic residues within the TMD of TA proteins better predict TA protein localization–the best consider just a five or seven residue face or a fraction of the TMD.
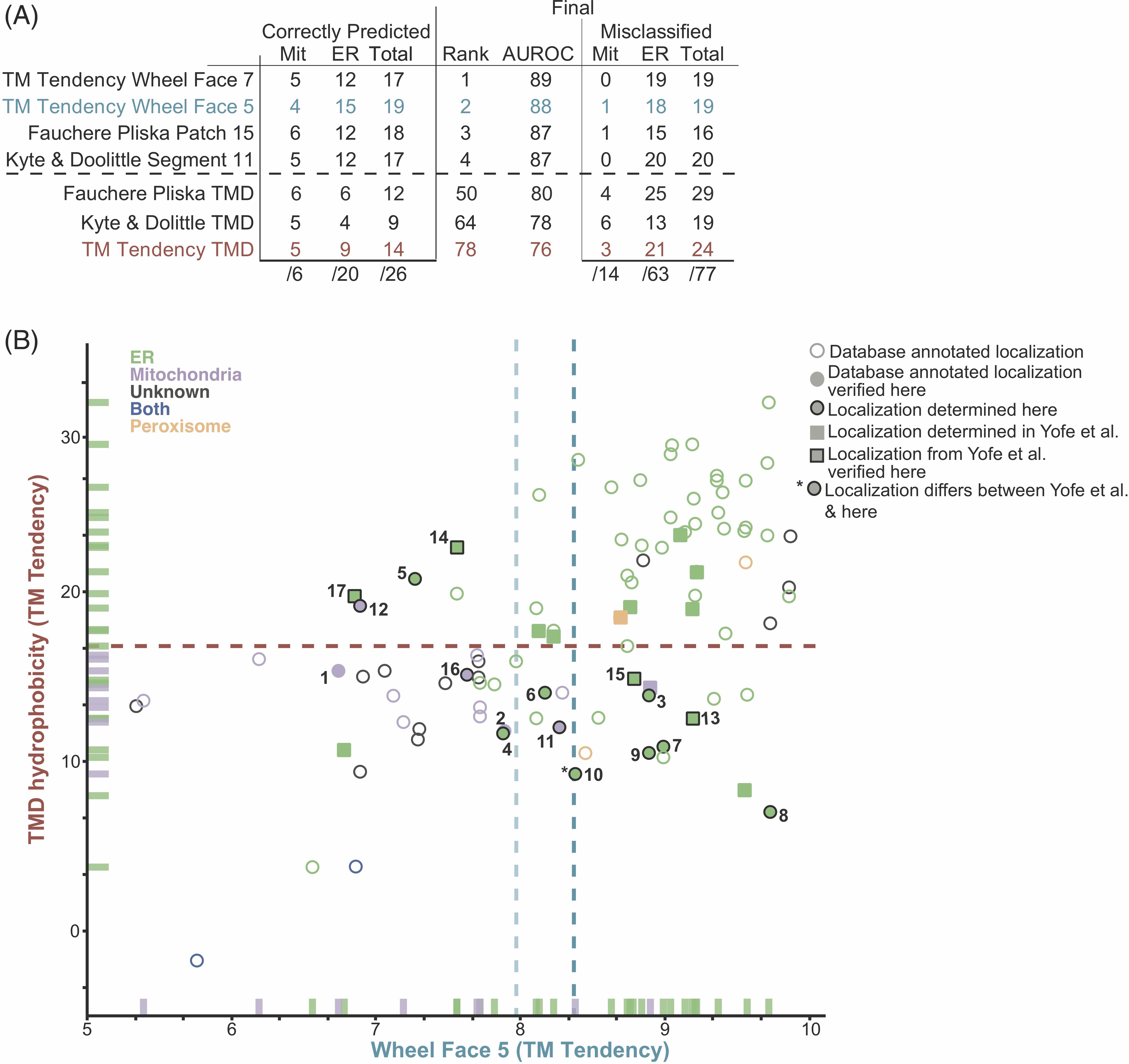
4.3.6 Expanding this metric to human TA proteins
We next applied this analysis to the human genome. Using our compiled list of 573 putative human TA proteins, we sought to identify a more inclusive set of criteria for ER- vs mitochondria-bound TA proteins. The best performing hydrophobicity scales in the yeast dataset were TM tendency and Kyte & Doolittle, so the other scales were not further considered with the human dataset. While TMD hydrophobicity metrics correctly capture mitochondria TA proteins, they fail to capture many ER TA proteins (Figure 4.6A, Table 4.7). Quantitatively assessing all metrics, we see slight improvements in classification with metrics using patches or segments compared to total hydrophobicity (Figure 4.6A,B; Table 4.7). The metric with the highest AUROC score is Patch 11 (Kyte and Doolittle). Many proteins in our dataset have a single report of their localizations in databases. There is potential for changes to these localizations as seen with many Bcl-2 family members (Figure 4.6B filled blue points) where there exist multiple reports of these proteins localizing to the ER and/or to the mitochondria. While this may be unique to these TA proteins, as their function to regulating apoptosis is tied in with their transport between the two membranes, some reported localizations may be the product of over-expression. Future work verifying and determining localizations of human TA proteins will likely result in improvements in classification by a metric derived from hydrophobic geometries.
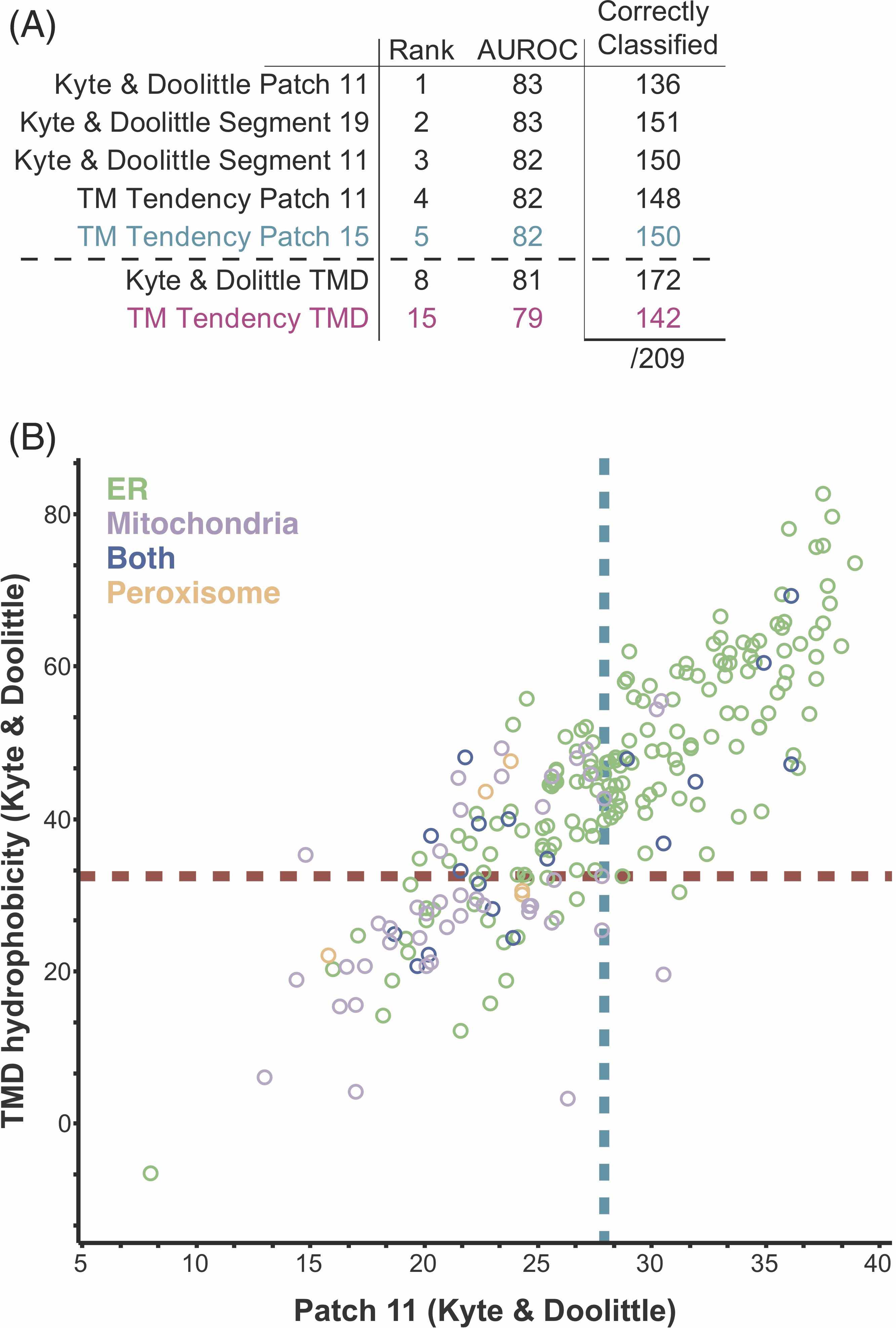
4.3.7 Determining a two-step criterion for localization determination
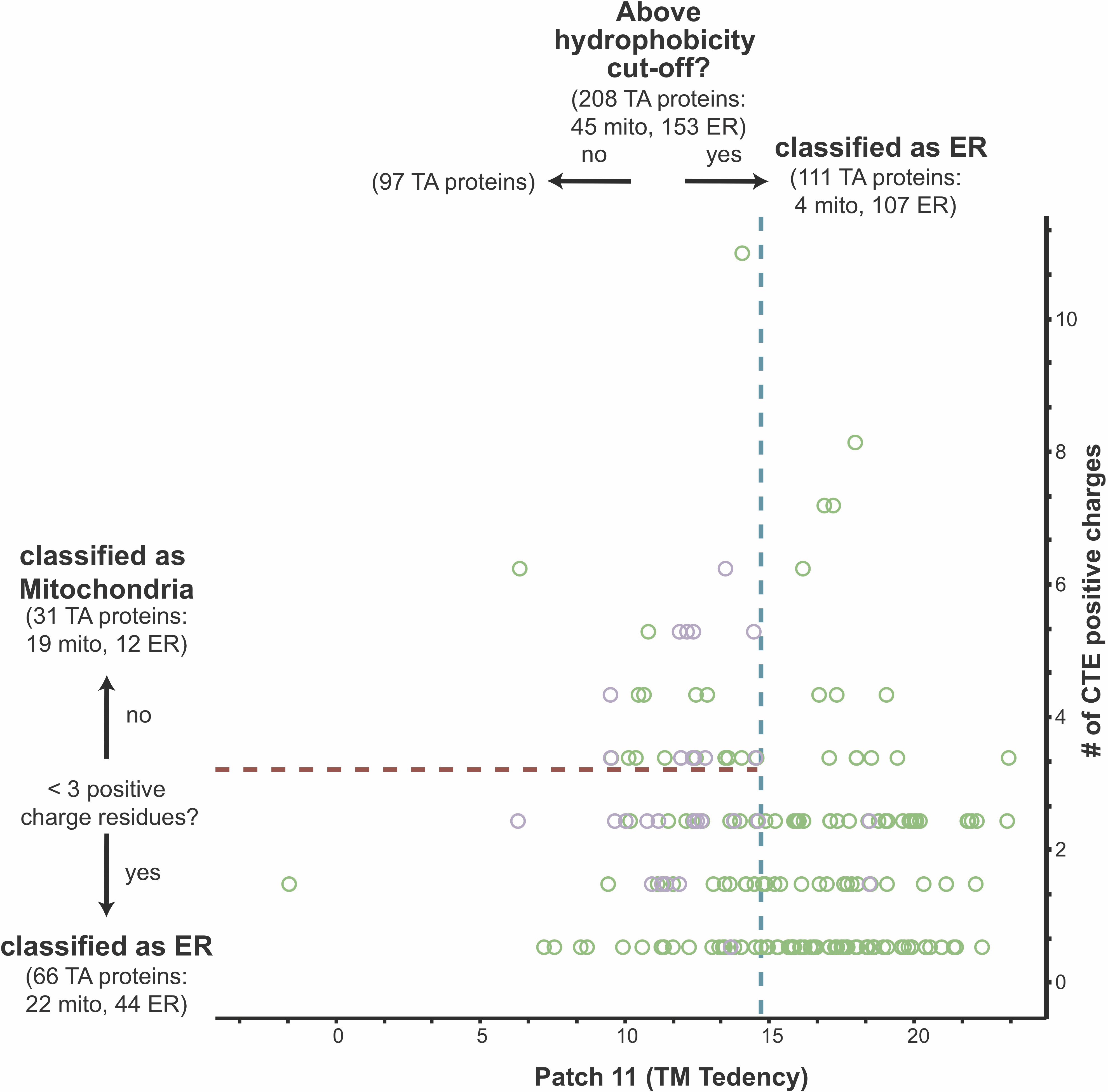
We then tested if combining a hydrophobicity geometry with a C-terminal charge metric resulted in more accurate classification of TA proteins. Costello and colleagues demonstrated in mammals, distinctions between ER, mitochondria and peroxisomal TA proteins can be made using a combination of charge and TMD hydrophobicity cut-offs (Costello et al., 2017). They suggest mitochondria TA proteins have tails that are less charged than peroxisomal TA proteins, but more charged than ER TA proteins, which are generally more hydrophobic than mitochondria TA proteins. Previous reports demonstrated the GET pathway fails to insert TA proteins with a sufficiently charged C-terminus (Rao et al., 2016). This selectivity filter was seen at the membrane and cytosolic components were unaffected by the presence of a charge. Perhaps this rejection of TA proteins with a C-terminal charge is seen across all ER targeting pathways in both yeast and humans. To further explore this, we determined anything to be above the hydrophobicity cut-off to be classified as ER-bound and anything below the cut-off to be passed through a charge filter. When analyzing the number of C-terminal positive residues following the TMD of TA proteins that fall below the hydrophobicity cut-off, we find that a benchmark of three positive residues best separates ER and mitochondria TA proteins–mitochondria TA proteins generally contain at least three charged residues. We applied this secondary filter to our best performing yeast metrics (Wheel Face 5 and Wheel Face 7 residues) and the TMD hydrophobicity (Table 4.1). In these cases, the three metrics perform the same, misclassifying 10 TA proteins. Intriguingly, a Patch 15 metric does best, correctly classifying 88% of all yeast TA proteins. A metric utilizing both a helical wheel face and C-terminal charge does slightly better than that using TMD hydrophobicity and charge, but the significance of that improvement is difficult to determine based on this small dataset.
The human dataset is larger, and we sought to apply this tandem metric application to our list of putative TA proteins (Figure 4.7). Similar to what was observed in the yeast dataset, improvements in classification are seen (Table 4.1). Interestingly, applying a C-terminal charge sequentially to hydrophobic metrics constrained to a fragment of ~11 residues, either a Patch (TM tendency) or the entire segment (Kyte & Doolittle), and the TMD hydrophobicity metric (Kyte & Doolittle), perform equally well, each misclassifying 38 TA proteins. Most hydrophobicity metrics performed similarly with either scale, suggesting a subset of the TMD is required for correct targeting (Table 4.1). It is clear that in both human and yeast, a combination of hydrophobicity and C-terminal charge filters are necessary for correct classification as was demonstrated in the context of the GET pathway. The hydrophobicity window can be limited to a fraction of the TMD and still perform as well as the entire TMD.
4.4 Discussion
Decoding the signaling information in membrane proteins responsible for their correct targeting to cellular membranes is still a mystery. For the class of membrane proteins with a single TMD and no signal peptide, TA proteins, some observations have been made to distinguish between those destined for the ER and those destined for the mitochondria. This report provides an extensive analysis of yeast and human TA proteins to identify a set of criteria to distinguish between ER- and mitochondria-bound TA proteins. This study also includes an expansion of putative TA proteins in both humans and yeast as well as newly determined experimental localization of several yeast TA proteins.
An initial separation by hydrophobicity can be applied to TA proteins, relegating TMDs with high hydrophobicities as ER proteins. A secondary filter can be applied to those below the cut-off classifying TA proteins with at least three charged residues following their TMDs as mitochondria-bound and the rest as ER-bound (Figure 4.7). This sequential selectivity was noted in the yeast GET pathway (Rao et al., 2016). In this case, it was demonstrated that the cytosolic targeting factors Sgt2 and Get3 bind to optimal TMDs based on a combination of high hydrophobicity and helical propensity. Regardless of hydrophobicity, TA proteins containing a charged C-termini were not inserted into ER microsomes. The analysis here demonstrates that generally ER TA proteins, not just GET substrates, lack charges in their C-terminus.
When determining the effectiveness of a hydrophobicity metric alone, metrics that focus on a hydrophobic geometry, a hydrophobic face in yeast and a hydrophobic segment restricted to 11 to 19 residues in humans, perform better than the hydrophobicity of the entire TMD. Applying the charge filter reveals that total hydrophobicity is as effective as hydrophobic face or segment metrics. Differences in the best performing hydrophobicity metrics between the yeast and human dataset could be explained by the observation that SGTA is more permissive to client binding than Sgt2 (Lin et al., 2021). Collectively, these datasets demonstrate that a fraction of the TMD is necessary and sufficient for correct localization. Interestingly, in the human dataset, some of the best performing metrics are limited to an 11-residue window, concurring with reports that SGTA recognizes TMDs of at least 11 amino acids (Lin et al., 2021).
While biochemical data suggested that clustering hydrophobic residues to one side of a helix increased binding to Sgt2, a co-chaperone in an ER TA protein targeting pathway, a cellular role of this hydrophobic face remained unclear (Lin et al., 2021). From the bioinformatic analysis and experimental localization data presented here, we demonstrate most yeast ER TA proteins contain a hydrophobic face–made of five to seven adjacent residues along a helical wheel plot. The two components of the GET pathways that directly bind to TA proteins, Sgt2 and Get3, both have binding sites composed of a hydrophobic groove. One could imagine the hydrophobic face in clients buried in the hydrophobic groove of Sgt2 and Get3, enhancing the hydrophobic binding interactions. Perhaps cellular factors involved in targeting TA proteins to the ER recognize this face and future identified ER TA protein binding partners will also feature a helical hand for client binding.
In this work, we provide a comprehensive bioinformatics analysis of naturally occurring TA proteins in the yeast and human genomes. While comprehensive, subtle differences in each metric’s geometries and hydrophobic scales cannot easily be differentiated analyzing just wild-type proteins. Similar work has helped disentangle the positional dependence of hydrophobicity in the insertion of integral membrane proteins (Hessa et al., 2007). Likewise, future work could better define the geometry and hydrophobic scale needed for TA targeting by larger scale mutational analyses, perhaps even transforming the question of TA targeting into that of sequence selection/enrichment (Fowler and Fields, 2014).
The targeting of TA proteins presents an intriguing and enigmatic problem for understanding the biogenesis of this important class of proteins. How subtle differences in clients modulate the interplay of hand-offs that direct these proteins to the correct membrane remains to be understood. Through in vivo imaging of yeast cells and computational analysis, we provide more clarity to client discrimination. A major outcome of this is the clear preference for a hydrophobic face on ER TA proteins of low hydrophobicity. In yeast, this alone is sufficient to predict the destination of a TA protein. In mammals, and likely more broadly in metazoans, while clearly an important component, alone the hydrophobic face cannot fully discriminate targets. For a full understanding, we expect other factors to contribute, reflective of the increased complexity of higher eukaryotes, perhaps involving more players (Aviram et al., 2016).
4.5 Material and Methods
4.5.1 Assembling a database of putative TA proteins and their TMDs
Proteins identified from UniProt (UniProt Consortium, 2021) containing a single TMD within 30 residues of the C-terminus were separated into groups based on their localization reported in UniProt. The topology of all proteins with 3 TMs or fewer was further analyzed using TOPCONS (Tsirigos et al., 2015) to avoid missed single-pass TM proteins. Proteins with a predicted signal peptide (Nielsen, 2017), an annotated transit peptide, problematic cautions, or with a length less than 50 or greater than 1000 residues were excluded. Proteins localized to the ER, golgi apparatus, nucleus, endosome, lysosome and cell membrane were classified as ER-bound, those localized to the outer mitochondrial membrane were classified as mitochondria-bound, those localized to the peroxisome were classified as peroxisomal proteins, and those with unknown localization were classified as unknown. Proteins with a compositional bias overlapping with the predicted TMD were also excluded. A handful of proteins and their inferred localizations were manually corrected or removed (see notebook and Table 4.2).
4.5.2 Assessing the predictive power of various hydrophobicity metrics
We thoroughly examined the metrics relating hydrophobicity, both published and by our own exploration, to better understand their relationship to protein localization. Notably, we recognized that a TMD’s hydrophobic moment (\(\mu\text{H}\)) (Eisenberg et al., 1984) was a poor predictor of localization, for example, although a Leu18 helix is extremely hydrophobic, it has (\(\mu\text{H}\)) = 0 since opposing hydrophobic residues are penalized in this metric. To address this, we define a metric that capture the presence of a hydrophobic face of the TMD: the maximally hydrophobic cluster on the face. For this metric, we sum the hydrophobicity of residues that orient sequentially on one side of a helix when visualized in a helical wheel diagram. While a range of hydrophobicity scales were predictive using this metric, we selected the TM Tendency scale (Zhao and London, 2006) to characterize the TMDs of putative TA proteins and determined the most predictive window by assessing a range of lengths from 4 to 12 (this would vary from three turns of a helix to six).
By considering sequences with inferred ER or mitochondrial localizations, we calculated the Area Under the Curve of a Receiver Operating Characteristic (AUROC) to assess predictive power. As we are comparing a real-valued metric (hydrophobicity) to a 2-class prediction, the AUROC is better suited for this analysis over others like accuracy or precision (a primer (Swets et al., 2000)). Because of many fewer mitochondrial proteins (ie, a class imbalance), we also confirmed that ordering hydrophobicity metrics by AUROC was consistent with the ordering produced by the more robust, but less common, Average Precision (see notebook).
4.5.3 Constructing plasmids for live cell imaging
A p416ADH-GFP-Fis1 plasmid and a mt-TagBFP described in Rao et al were gifted to us from the Walter lab, UCSF (Okreglak and Walter, 2014; Rao et al., 2016) and a Sec63-tdtomato was a gift from Sebastian Schuck, ZMVH, Universitat Heidelberg. TMDs sequences were ordered from Twist Biosciences (San Francisco, CA) with flanking HindIII and XhoI sites. GFP-TMD constructs were made by restriction enzyme digestion (New England Biolabs, USA) of the p416ADH-GFP-Fis1 plasmid and the genes ordered from Twist Biosciences followed by T4 DNA (New England Biolabs, USA) ligation of the template and TMD fragments.
4.5.4 Live cell imaging
The yeast strain used are those described in Rao et al, also a gift from the Walter Lab, UCSF. Strains containing each GFP fused TMD were grown in appropriate selection media. Coverslips were prepped by coating with 0.1 mg/mL concavalin A (Sigma, USA) in 0.9% NaCl solution. Cells were immobilized on coverslips at a concentration of 5000 cells/mm2 (plates at 1.8 cm2, thus 9 × 108 cells/well) and imaged using a Nikon LSM800 (Nikon, Japan). Images were collected at wavelengths 488, 514 and 581 nm and were processed with ImageJ (Schneider et al., 2012) and two in-house image processing algorithms.
4.5.5 Image processing to determine localization
Yeast cells were segmented using deep learning-based tools. The variable pattern of DIC images with mixed low and high contrasts for back-grounds and cell bodies (signal variance of each whole image ranging from 67.4 to 2706.3, a ×40 difference–average, median and SD of signal variance for all images were, respectively 645.6, 563.8 and 419.1) prevented using classical gradient based methods to successfully segment cells. We adopted and compared two contemporary tools, YeastSpotter, a Mask-RCNN method dedicated to yeast cells (Lu et al., 2019), and Cellpose, a generalist method trained on a large pool of cell images (Stringer et al., 2021). Note that, the former was not trained on yeast cell images but used a model pretrained on a larger set of other cell images to build a friendly tool for yeast cell segmentation. Cellpose is a more sophisticated tool whose pretrained models have learned to segment well based on a myriad of intensity gradient values and image styles. It has shown to achieve high quality segmentation on an extended variety of cell images, including in our yeast cells images, producing superior results when compared to YeastSpotter with the advantage of running faster on GPUs (tested on Nvidia RTX 2080 Ti). We thus exclusively used Cellpose with its cyto pretrained model to segment yeast cells in all our DIC images. We used maximum intensity projections of up to two or three slices per image stack but mostly a single slice was sufficient to create a single representative image for segmentation. Spurious, tiny, segmented regions whose size were shown to be outliers were automatically removed using an area opening morphological operation.
Individual cells were isolated by applying the mask to the corresponding florescent images of each of the three wavelengths. Masks less than \(7.5 \mu\text{m}^2\) corresponded to incorrectly identified, incomplete, or out-of-plane cells and were omitted from analysis. Masks were applied to each florescence channel. An empirical threshold was applied to each channel to identify true florescence from background, and the percentage of each cell with co-localized GFP and BFP or GFP and tdTomato was then calculated. Localization was then determined identifying which pair of channels (GFP&BFP vs GFP&tdTomato) had greater overlap, that is, OverlapGFP&BFP > OverlapGFP&tdTomato resulted in a mitochondria annotation. The number of individual cells in each category were counted. Outputs from this algorithm were verified by manually inspecting individual images.
4.5.6 Data Availability
All code employed is available openly at http://github.com/clemlab/sgt2a-modeling with analysis done in Jupyter Lab/Notebooks using Python 3.6 enabled by Numpy, Pandas, Scikit-Learn, BioPython, bebi103 (Bois, 2020), and Bokeh as well as in Rstudio/Rmarkdown Notebooks enabled by packages within the Tidyverse ecosystem.
4.6 Supplementary Figure
4.7 Supplementary Tables
| i | Organism | Entry | Gene names | Localization |
|---|---|---|---|---|
| 1 | H. sapiens | Q16611 | BAK1 BAK BCL2L7 CDN1 | Both |
| 2 | H. sapiens | P10415 | BCL2 | Both |
| 3 | H. sapiens | O60238 | BNIP3L BNIP3A BNIP3H NIX | Both |
| 4 | H. sapiens | P26678 | PLN PLB | Both |
| 5 | H. sapiens | Q12981 | BNIP1 NIP1 SEC20L TRG8 | Both |
| 6 | H. sapiens | Q09013 | DMPK DM1PK MDPK | Both |
| 7 | H. sapiens | Q969F0 | FATE1 FATE | Both |
| 8 | H. sapiens | Q07812 | BAX BCL2L4 | Both |
| 9 | H. sapiens | Q07817 | BCL2L1 BCL2L BCLX | Both |
| 10 | H. sapiens | Q9HD36 | BCL2L10 BCLB | Both |
| 11 | H. sapiens | Q12983 | BNIP3 NIP3 | Both |
| 12 | H. sapiens | Q9UMX3 | BOK BCL2L9 | Both |
| 13 | H. sapiens | Q9BWH2 | FUNDC2 HCBP6 DC44 HCC3 PD03104 | Both |
| 14 | H. sapiens | Q07820 | MCL1 BCL2L3 | Both |
| 15 | H. sapiens | Q86Y07 | VRK2 | Both |
| 16 | H. sapiens | P23763 | VAMP1 SYB1 | Both |
| 17 | H. sapiens | A0A0A0MTJ1 | FKBP8 | Both |
| 18 | H. sapiens | I3L3X5 | PLSCR3 | Both |
| 19 | H. sapiens | P18031 | PTPN1 PTP1B | ER |
| 20 | H. sapiens | Q13323 | BIK NBK | ER |
| 21 | H. sapiens | Q9H305 | CDIP1 C16orf5 CDIP LITAFL | ER |
| 22 | H. sapiens | Q5VV42 | CDKAL1 | ER |
| 23 | H. sapiens | A4D256 | CDC14C CDC14B2 CDC14Bretro | ER |
| 24 | H. sapiens | Q96JN2 | CCDC136 KIAA1793 NAG6 | ER |
| 25 | H. sapiens | Q9NXE4 | SMPD4 KIAA1418 SKNY | ER |
| 26 | H. sapiens | Q9H0X9 | OSBPL5 KIAA1534 OBPH1 ORP5 | ER |
| 27 | H. sapiens | Q9BZF1 | OSBPL8 KIAA1451 ORP8 OSBP10 | ER |
| 28 | H. sapiens | Q9NZM1 | MYOF FER1L3 KIAA1207 | ER |
| 29 | H. sapiens | Q9HC10 | OTOF FER1L2 | ER |
| 30 | H. sapiens | Q9HCU5 | PREB SEC12 | ER |
| 31 | H. sapiens | O15162 | PLSCR1 | ER |
| 32 | H. sapiens | Q9NRQ2 | PLSCR4 GIG43 | ER |
| 33 | H. sapiens | A0PG75 | PLSCR5 | ER |
| 34 | H. sapiens | Q9NRY7 | PLSCR2 | ER |
| 35 | H. sapiens | P50876 | RNF144A KIAA0161 RNF144 UBCE7IP4 | ER |
| 36 | H. sapiens | Q9NZ42 | PSENEN PEN2 MDS033 | ER |
| 37 | H. sapiens | Q96CS7 | PLEKHB2 EVT2 | ER |
| 38 | H. sapiens | Q6ZNB6 | NFXL1 OZFP | ER |
| 39 | H. sapiens | P17706 | PTPN2 PTPT | ER |
| 40 | H. sapiens | Q7Z6L0 | PRRT2 | ER |
| 41 | H. sapiens | Q9Y6X1 | SERP1 RAMP4 | ER |
| 42 | H. sapiens | P61266 | STX1B STX1B1 STX1B2 | ER |
| 43 | H. sapiens | Q9UNK0 | STX8 | ER |
| 44 | H. sapiens | Q16623 | STX1A STX1 | ER |
| 45 | H. sapiens | Q8WXE9 | STON2 STN2 STNB | ER |
| 46 | H. sapiens | Q86Y82 | STX12 | ER |
| 47 | H. sapiens | Q7Z699 | SPRED1 | ER |
| 48 | H. sapiens | Q9P2W9 | STX18 GIG9 | ER |
| 49 | H. sapiens | Q13190 | STX5 STX5A | ER |
| 50 | H. sapiens | O15400 | STX7 | ER |
| 51 | H. sapiens | P32856 | STX2 EPIM STX2A STX2B STX2C | ER |
| 52 | H. sapiens | O60499 | STX10 SYN10 | ER |
| 53 | H. sapiens | Q13277 | STX3 STX3A | ER |
| 54 | H. sapiens | O14662 | STX16 | ER |
| 55 | H. sapiens | Q5QGT7 | RTP2 Z3CXXC2 | ER |
| 56 | H. sapiens | P59025 | RTP1 Z3CXXC1 | ER |
| 57 | H. sapiens | Q9BQQ7 | RTP3 TMEM7 Z3CXXC3 | ER |
| 58 | H. sapiens | Q8N205 | SYNE4 C19orf46 | ER |
| 59 | H. sapiens | Q6ZMZ3 | SYNE3 C14orf139 C14orf49 LINC00341 | ER |
| 60 | H. sapiens | B2RUZ4 | SMIM1 | ER |
| 61 | H. sapiens | Q7Z698 | SPRED2 | ER |
| 62 | H. sapiens | Q96DX8 | RTP4 IFRG28 Z3CXXC4 | ER |
| 63 | H. sapiens | Q96QK8 | SMIM14 C4orf34 | ER |
| 64 | H. sapiens | Q14BN4 | SLMAP KIAA1601 SLAP UNQ1847/PRO3577 | ER |
| 65 | H. sapiens | Q86T96 | RNF180 | ER |
| 66 | H. sapiens | Q8N8N0 | RNF152 | ER |
| 67 | H. sapiens | Q12846 | STX4 STX4A | ER |
| 68 | H. sapiens | O43752 | STX6 | ER |
| 69 | H. sapiens | P60059 | SEC61G | ER |
| 70 | H. sapiens | Q14D33 | RTP5 C2orf85 CXXC11 Z3CXXC5 | ER |
| 71 | H. sapiens | P60468 | SEC61B | ER |
| 72 | H. sapiens | Q8N6R1 | SERP2 C13orf21 | ER |
| 73 | H. sapiens | P01850 | TRBC1 | ER |
| 74 | H. sapiens | P03986 | TRGC2 TCRGC2 | ER |
| 75 | H. sapiens | Q629K1 | TRIQK C8orf83 | ER |
| 76 | H. sapiens | A0A5B9 | TRBC2 TCRBC2 | ER |
| 77 | H. sapiens | Q9NSU2 | TREX1 | ER |
| 78 | H. sapiens | P01848 | TRAC TCRA | ER |
| 79 | H. sapiens | B7Z8K6 | TRDC | ER |
| 80 | H. sapiens | Q96D59 | RNF183 | ER |
| 81 | H. sapiens | P00167 | CYB5A CYB5 | ER |
| 82 | H. sapiens | O75923 | DYSF FER1L1 | ER |
| 83 | H. sapiens | P50402 | EMD EDMD STA | ER |
| 84 | H. sapiens | O42043 | ERVK-18 | ER |
| 85 | H. sapiens | Q52LJ0 | FAM98B | ER |
| 86 | H. sapiens | Q9NYM9 | BET1L GS15 | ER |
| 87 | H. sapiens | P54710 | FXYD2 ATP1C ATP1G1 | ER |
| 88 | H. sapiens | O95415 | BRI3 | ER |
| 89 | H. sapiens | Q9BXU9 | CALN1 CABP8 | ER |
| 90 | H. sapiens | O15155 | BET1 | ER |
| 91 | H. sapiens | Q86V35 | CABP7 CALN2 | ER |
| 92 | H. sapiens | Q01740 | FMO1 | ER |
| 93 | H. sapiens | P49326 | FMO5 | ER |
| 94 | H. sapiens | P31513 | FMO3 | ER |
| 95 | H. sapiens | Q8N8J7 | FAM241A C4orf32 | ER |
| 96 | H. sapiens | P31512 | FMO4 FMO2 | ER |
| 97 | H. sapiens | Q9P0K9 | FRRS1L C9orf4 | ER |
| 98 | H. sapiens | Q9Y2H6 | FNDC3A FNDC3 HUGO KIAA0970 | ER |
| 99 | H. sapiens | P13164 | IFITM1 CD225 IFI17 | ER |
| 100 | H. sapiens | Q01629 | IFITM2 | ER |
| 101 | H. sapiens | Q01628 | IFITM3 | ER |
| 102 | H. sapiens | Q8TBA6 | GOLGA5 RETII RFG5 PIG31 | ER |
| 103 | H. sapiens | O95249 | GOSR1 GS28 | ER |
| 104 | H. sapiens | Q14789 | GOLGB1 | ER |
| 105 | H. sapiens | Q96JJ6 | JPH4 JPHL1 KIAA1831 | ER |
| 106 | H. sapiens | Q9HDC5 | JPH1 JP1 | ER |
| 107 | H. sapiens | Q9BR39 | JPH2 JP2 | ER |
| 108 | H. sapiens | Q8WXH2 | JPH3 JP3 TNRC22 | ER |
| 109 | H. sapiens | Q99732 | LITAF PIG7 SIMPLE | ER |
| 110 | H. sapiens | O75427 | LRCH4 LRN LRRN1 LRRN4 | ER |
| 111 | H. sapiens | Q9Y2L9 | LRCH1 CHDC1 KIAA1016 | ER |
| 112 | H. sapiens | Q3KP22 | MAJIN C11orf85 | ER |
| 113 | H. sapiens | Q86Z14 | KLB | ER |
| 114 | H. sapiens | Q9Y6H6 | KCNE3 | ER |
| 115 | H. sapiens | P42167 | TMPO LAP2 | ER |
| 116 | H. sapiens | P30519 | HMOX2 HO2 | ER |
| 117 | H. sapiens | Q8WWP7 | GIMAP1 IMAP1 | ER |
| 118 | H. sapiens | Q96F15 | GIMAP5 IAN4L1 IAN5 IMAP3 | ER |
| 119 | H. sapiens | P09601 | HMOX1 HO HO1 | ER |
| 120 | H. sapiens | Q9UPX6 | MINAR1 KIAA1024 | ER |
| 121 | H. sapiens | Q8NHP6 | MOSPD2 | ER |
| 122 | H. sapiens | P51648 | ALDH3A2 ALDH10 FALDH | ER |
| 123 | H. sapiens | Q8N2K1 | UBE2J2 NCUBE2 | ER |
| 124 | H. sapiens | O94966 | USP19 KIAA0891 ZMYND9 | ER |
| 125 | H. sapiens | Q9NZ43 | USE1 USE1L MDS032 | ER |
| 126 | H. sapiens | Q9P0L0 | VAPA VAP33 | ER |
| 127 | H. sapiens | O95159 | ZFPL1 | ER |
| 128 | H. sapiens | Q5T7W0 | ZNF618 KIAA1952 | ER |
| 129 | H. sapiens | O14653 | GOSR2 GS27 | ER |
| 130 | H. sapiens | P51809 | VAMP7 SYBL1 | ER |
| 131 | H. sapiens | Q9BV40 | VAMP8 | ER |
| 132 | H. sapiens | Q9UEU0 | VTI1B VTI1 VTI1L VTI1L1 VTI2 | ER |
| 133 | H. sapiens | Q96AJ9 | VTI1A | ER |
| 134 | H. sapiens | O95292 | VAPB UNQ484/PRO983 | ER |
| 135 | H. sapiens | O95183 | VAMP5 HSPC191 | ER |
| 136 | H. sapiens | Q15836 | VAMP3 SYB3 | ER |
| 137 | H. sapiens | O75379 | VAMP4 | ER |
| 138 | H. sapiens | Q9Y385 | UBE2J1 NCUBE1 CGI-76 HSPC153 HSPC205 | ER |
| 139 | H. sapiens | P63027 | VAMP2 SYB2 | ER |
| 140 | H. sapiens | A0A1W2PPG1 | GOSR2 | ER |
| 141 | H. sapiens | A0A087WWT2 | NRN1 | ER |
| 142 | H. sapiens | E9PLT1 | CD36 | ER |
| 143 | H. sapiens | E7ENI6 | ICA1 | ER |
| 144 | H. sapiens | E9PN33 | STX3 | ER |
| 145 | H. sapiens | X6R383 | SETDB2 | ER |
| 146 | H. sapiens | K7EJC8 | GOSR1 | ER |
| 147 | H. sapiens | A0A1W2PRH6 | PAX6 | ER |
| 148 | H. sapiens | D6RE10 | ELOVL7 | ER |
| 149 | H. sapiens | F5H2S3 | P2RX4 | ER |
| 150 | H. sapiens | F8WAT4 | PAPOLG | ER |
| 151 | H. sapiens | F5H3K6 | SPI1 | ER |
| 152 | H. sapiens | E9PE96 | PCLO | ER |
| 153 | H. sapiens | D6RF86 | CDH6 | ER |
| 154 | H. sapiens | E7ETP9 | LAMP3 | ER |
| 155 | H. sapiens | A0A1W2PRF6 | SCARB2 | ER |
| 156 | H. sapiens | B4DSN5 | PTPN1 | ER |
| 157 | H. sapiens | D6RBD7 | EEF1E1 hCG_15559 | ER |
| 158 | H. sapiens | A0A087WTJ2 | GIMAP1-GIMAP5 | ER |
| 159 | H. sapiens | A0A1W2PS81 | GOSR2 | ER |
| 160 | H. sapiens | A0A087WWT0 | JPH4 | ER |
| 161 | H. sapiens | A0A0J9YW33 | STX3 | ER |
| 162 | H. sapiens | C9JUH5 | SERP1 | ER |
| 163 | H. sapiens | H7C410 | GPC1 | ER |
| 164 | H. sapiens | U3KQS5 | TATDN1 | ER |
| 165 | H. sapiens | G5EA09 | SDCBP hCG_1787561 | ER |
| 166 | H. sapiens | B1AL79 | PKN2 hCG_23733 | ER |
| 167 | H. sapiens | B7Z5N5 | SMAD2 | ER |
| 168 | H. sapiens | I3L3H3 | P2RX1 | ER |
| 169 | H. sapiens | A0A087WT82 | GPC6 | ER |
| 170 | H. sapiens | F5H895 | DAD1 | ER |
| 171 | H. sapiens | F2Z2S5 | SERP2 | ER |
| 172 | H. sapiens | A0A1B0GTF8 | EPB41L3 | ER |
| 173 | H. sapiens | E9PCT3 | CAV2 | ER |
| 174 | H. sapiens | F8WBE5 | TFRC | ER |
| 175 | H. sapiens | K7EQG9 | PTPN2 | ER |
| 176 | H. sapiens | A0A0U1RQC9 | TP53 | ER |
| 177 | H. sapiens | F8WCE5 | LMLN | ER |
| 178 | H. sapiens | A0SDD8 | CLDN16 | ER |
| 179 | H. sapiens | B5MCA4 | EPCAM | ER |
| 180 | H. sapiens | Q5HY57 | EMD hCG_41343 | ER |
| 181 | H. sapiens | B4DJ94 | ATP9B | ER |
| 182 | H. sapiens | Q86XC5 | TMEM97 | ER |
| 183 | H. sapiens | Q9NRY6 | PLSCR3 | Mit |
| 184 | H. sapiens | Q7Z419 | RNF144B IBRDC2 P53RFP | Mit |
| 185 | H. sapiens | P56378 | MP68 C14orf2 PRO1574 | Mit |
| 186 | H. sapiens | P57105 | SYNJ2BP OMP25 | Mit |
| 187 | H. sapiens | Q9P0U1 | TOMM7 TOM7 TOMM07 AD-014 | Mit |
| 188 | H. sapiens | Q8N4H5 | TOMM5 C9orf105 TOM5 | Mit |
| 189 | H. sapiens | Q8WWH4 | ASZ1 ALP1 ANKL1 C7orf7 GASZ | Mit |
| 190 | H. sapiens | P27338 | MAOB | Mit |
| 191 | H. sapiens | Q9BXK5 | BCL2L13 MIL1 CD003 | Mit |
| 192 | H. sapiens | O43169 | CYB5B CYB5M OMB5 | Mit |
| 193 | H. sapiens | Q14318 | FKBP8 FKBP38 | Mit |
| 194 | H. sapiens | Q9Y3D6 | FIS1 TTC11 CGI-135 | Mit |
| 195 | H. sapiens | Q96I36 | COX14 C12orf62 | Mit |
| 196 | H. sapiens | O00198 | HRK BID3 | Mit |
| 197 | H. sapiens | Q14410 | GK2 GKP2 GKTA | Mit |
| 198 | H. sapiens | Q9GZY8 | MFF C2orf33 AD030 AD033 GL004 | Mit |
| 199 | H. sapiens | Q7Z434 | MAVS IPS1 KIAA1271 VISA | Mit |
| 200 | H. sapiens | Q8IXI1 | RHOT2 ARHT2 C16orf39 | Mit |
| 201 | H. sapiens | Q13505 | MTX1 MTX MTXN | Mit |
| 202 | H. sapiens | Q8IXI2 | RHOT1 ARHT1 | Mit |
| 203 | H. sapiens | P21397 | MAOA | Mit |
| 204 | H. sapiens | Q14409 | GK3P GKP3 GKTB | Mit |
| 205 | H. sapiens | Q96IX5 | USMG5 DAPIT HCVFTP2 PD04912 | Mit |
| 206 | H. sapiens | A0A087WT64 | MCL1 | Mit |
| 207 | H. sapiens | E9PH05 | FAM162A | Mit |
| 208 | H. sapiens | A0A0C4DFQ1 | MTX1 | Mit |
| 209 | H. sapiens | A0A087WZY2 | TOMM7 | Mit |
| 210 | H. sapiens | C9JU26 | ATP5MF | Mit |
| 211 | H. sapiens | S4R2X2 | SFXN1 | Mit |
| 212 | H. sapiens | H7BXZ6 | RHOT1 hCG_1991479 | Mit |
| 213 | H. sapiens | A0A0A0MS29 | MFF | Mit |
| 214 | H. sapiens | J3KNF8 | CYB5B hCG_28205 | Mit |
| 215 | H. sapiens | P56134 | ATP5J2 ATP5JL | MitIn |
| 216 | H. sapiens | O43676 | NDUFB3 | MitIn |
| 217 | H. sapiens | O14957 | UQCR11 UQCR | MitIn |
| 218 | H. sapiens | Q9UDW1 | UQCR10 UCRC HSPC119 | MitIn |
| 219 | H. sapiens | O95168 | NDUFB4 | MitIn |
| 220 | H. sapiens | P09669 | COX6C | MitIn |
| 221 | H. sapiens | Q9Y2R0 | COA3 CCDC56 MITRAC12 HSPC009 | MitIn |
| 222 | H. sapiens | Q96AQ8 | MCUR1 C6orf79 CCDC90A | MitIn |
| 223 | H. sapiens | F8WAR4 | CHCHD3 | MitIn |
| 224 | H. sapiens | D6R9C3 | COX7A2 | MitIn |
| 225 | H. sapiens | A0A087WU07 | MINOS1 | MitIn |
| 226 | H. sapiens | A0A087WYS9 | SURF1 | MitIn |
| 227 | H. sapiens | C9IZW8 | NDUFB2 | MitIn |
| 228 | H. sapiens | O96011 | PEX11B | Pex |
| 229 | H. sapiens | Q8NFP0 | PXT1 STEPP | Pex |
| 230 | H. sapiens | P53816 | PLA2G16 HRASLS3 HREV107 | Pex |
| 231 | H. sapiens | Q5T8D3 | ACBD5 KIAA1996 | Pex |
| 232 | H. sapiens | B7Z2R7 | ACBD5 | Pex |
| 233 | H. sapiens | Q9P0B6 | CCDC167 C6orf129 HSPC265 | Unknown |
| 234 | H. sapiens | Q8N111 | CEND1 BM88 | Unknown |
| 235 | H. sapiens | Q8WVX3 | C4orf3 | Unknown |
| 236 | H. sapiens | Q9H7X2 | C1orf115 | Unknown |
| 237 | H. sapiens | Q6ZSY5 | PPP1R3F | Unknown |
| 238 | H. sapiens | Q9UF11 | PLEKHB1 EVT1 KPL1 PHR1 PHRET1 | Unknown |
| 239 | H. sapiens | Q6ZS82 | RGS9BP R9AP | Unknown |
| 240 | H. sapiens | Q16821 | PPP1R3A PP1G | Unknown |
| 241 | H. sapiens | Q9NS64 | RPRM | Unknown |
| 242 | H. sapiens | Q6IEE8 | SLFN12L SLFN5 | Unknown |
| 243 | H. sapiens | Q9NRQ5 | SMCO4 C11orf75 FN5 | Unknown |
| 244 | H. sapiens | Q8NCU8 | SMIM37 LINC00116 NCRNA00116 | Unknown |
| 245 | H. sapiens | Q96HG1 | SMIM10 CXorf69 | Unknown |
| 246 | H. sapiens | Q71RC9 | SMIM5 C17orf109 | Unknown |
| 247 | H. sapiens | P0DL12 | SMIM17 | Unknown |
| 248 | H. sapiens | Q8WVI0 | SMIM4 C3orf78 | Unknown |
| 249 | H. sapiens | A0A1B0GUA5 | SMIM32 | Unknown |
| 250 | H. sapiens | Q96KF7 | SMIM8 C6orf162 DC18 | Unknown |
| 251 | H. sapiens | Q8TC41 | RNF217 C6orf172 IBRDC1 | Unknown |
| 252 | H. sapiens | Q9Y228 | TRAF3IP3 T3JAM | Unknown |
| 253 | H. sapiens | Q5JXX7 | TMEM31 | Unknown |
| 254 | H. sapiens | Q2MJR0 | SPRED3 EVE-3 | Unknown |
| 255 | H. sapiens | L0R6Q1 | SLC35A4 | Unknown |
| 256 | H. sapiens | O75920 | SERF1A FAM2A SERF1 SMAM1; SERF1B FAM2B SERF1 SMAM1 | Unknown |
| 257 | H. sapiens | Q9H4I3 | TRABD TTG2 PP2447 | Unknown |
| 258 | H. sapiens | A6NCQ9 | RNF222 | Unknown |
| 259 | H. sapiens | Q5SWX8 | ODR4 C1orf27 TTG1 TTG1A | Unknown |
| 260 | H. sapiens | Q8N326 | C10orf111 | Unknown |
| 261 | H. sapiens | Q96LL3 | C16orf92 | Unknown |
| 262 | H. sapiens | Q6P4D5 | FAM122C | Unknown |
| 263 | H. sapiens | Q96D05 | FAM241B C10orf35 | Unknown |
| 264 | H. sapiens | Q8N7S6 | ARIH2OS C3orf71 | Unknown |
| 265 | H. sapiens | Q8IVJ8 | APRG1 C3orf35 | Unknown |
| 266 | H. sapiens | Q86W74 | ANKRD46 | Unknown |
| 267 | H. sapiens | Q8WVC6 | DCAKD | Unknown |
| 268 | H. sapiens | Q2WGJ9 | FER1L6 C8orfK23 | Unknown |
| 269 | H. sapiens | Q5RGS3 | FAM74A1 | Unknown |
| 270 | H. sapiens | A9Z1Z3 | FER1L4 C20orf124 | Unknown |
| 271 | H. sapiens | Q53EP0 | FNDC3B FAD104 NS5ABP37 UNQ2421/PRO4979/PRO34274 | Unknown |
| 272 | H. sapiens | Q96JQ2 | CLMN KIAA1188 | Unknown |
| 273 | H. sapiens | Q9NPU4 | C14orf132 C14orf88 | Unknown |
| 274 | H. sapiens | Q6ZS62 | COLCA1 C11orf92 | Unknown |
| 275 | H. sapiens | A1L1A6 | IGSF23 | Unknown |
| 276 | H. sapiens | Q8IUY3 | GRAMD2A GRAMD2 | Unknown |
| 277 | H. sapiens | Q9NWW9 | HRASLS2 | Unknown |
| 278 | H. sapiens | Q9UL19 | RARRES3 RIG1 TIG3 | Unknown |
| 279 | H. sapiens | Q68G75 | LEMD1 | Unknown |
| 280 | H. sapiens | P59773 | KIAA1024L | Unknown |
| 281 | H. sapiens | Q9HDD0 | HRASLS | Unknown |
| 282 | H. sapiens | Q96EZ4 | MYEOV OCIM | Unknown |
| 283 | H. sapiens | A0A024RCL3 | MICA hCG_2001511 | Unknown |
| 284 | H. sapiens | A0A087WXU0 | RMND1 | Unknown |
| 285 | H. sapiens | I3L1J9 | TNFRSF12A | Unknown |
| 286 | H. sapiens | E9PLR7 | RNF121 | Unknown |
| 287 | H. sapiens | H0YIU3 | RNASEK | Unknown |
| 288 | H. sapiens | E7EX18 | MPV17 | Unknown |
| 289 | H. sapiens | H0YNW0 | SLC12A1 | Unknown |
| 290 | H. sapiens | A0A087WWM7 | MME | Unknown |
| 291 | H. sapiens | F5GZV7 | VAMP1 | Unknown |
| 292 | H. sapiens | A0A0D9SGD9 | SLFN12L | Unknown |
| 293 | H. sapiens | J3KR13 | FOLR2 | Unknown |
| 294 | H. sapiens | A0A087X240 | EFNA5 | Unknown |
| 295 | H. sapiens | A0A1W2PRR9 | EGFR | Unknown |
| 296 | H. sapiens | C9JD05 | FSD1L | Unknown |
| 297 | H. sapiens | C9JXZ5 | VAMP8 | Unknown |
| 298 | H. sapiens | E9PQR3 | FTH1 | Unknown |
| 299 | H. sapiens | J3KNC7 | CYB5A | Unknown |
| 300 | H. sapiens | J3KPI8 | GPR139 | Unknown |
| 301 | H. sapiens | E9PQY3 | ACP2 | Unknown |
| 302 | H. sapiens | E7EPM7 | COQ2 | Unknown |
| 303 | H. sapiens | A0A087X286 | CKLF-CMTM1 | Unknown |
| 304 | H. sapiens | V9GYT2 | ANKRD29 | Unknown |
| 305 | H. sapiens | K7EQB1 | STX8 | Unknown |
| 306 | H. sapiens | E9PAR0 | FKBP11 | Unknown |
| 307 | H. sapiens | J3QS48 | MPDU1 | Unknown |
| 308 | H. sapiens | H7BXF4 | SMPD4 | Unknown |
| 309 | H. sapiens | F8WDY4 | TMBIM1 | Unknown |
| 310 | H. sapiens | J3KN43 | TMEM33 | Unknown |
| 311 | H. sapiens | A8MTT8 | ZNF286A | Unknown |
| 312 | H. sapiens | C9JYK0 | LRCH4 | Unknown |
| 313 | H. sapiens | E5RFY6 | RNF217 | Unknown |
| 314 | H. sapiens | E9PFI9 | ZP3 | Unknown |
| 315 | H. sapiens | A0A0J9YWK4 | HBB | Unknown |
| 316 | H. sapiens | A0A0D9SFF9 | MELK | Unknown |
| 317 | H. sapiens | A0A1W2PPL1 | SPIN3 | Unknown |
| 318 | H. sapiens | F8WF90 | ARL6IP5 | Unknown |
| 319 | H. sapiens | F5H0W1 | DPY19L2 | Unknown |
| 320 | H. sapiens | F5H543 | IYD | Unknown |
| 321 | H. sapiens | A0A1W2PRW4 | PLA2G16 | Unknown |
| 322 | H. sapiens | E9PKL4 | C11orf96 | Unknown |
| 323 | H. sapiens | E9PNH0 | OSBPL5 | Unknown |
| 324 | H. sapiens | E9PJ90 | HBS1L | Unknown |
| 325 | H. sapiens | E9PPZ2 | NPEPPS | Unknown |
| 326 | H. sapiens | C9IZ55 | MALL | Unknown |
| 327 | H. sapiens | A8MPV4 | MPV17 | Unknown |
| 328 | H. sapiens | H0YNT6 | FES | Unknown |
| 329 | H. sapiens | F5H1L9 | JPH4 | Unknown |
| 330 | H. sapiens | A0A087X175 | SLC38A3 | Unknown |
| 331 | H. sapiens | A0A286YEN9 | C5orf60 | Unknown |
| 332 | H. sapiens | A0A1W2PP90 | ST3GAL5 | Unknown |
| 333 | H. sapiens | H3BUG9 | TMEM202 | Unknown |
| 334 | H. sapiens | A0A1W2PQZ3 | HLA-B | Unknown |
| 335 | H. sapiens | D6R9K1 | CLDND1 | Unknown |
| 336 | H. sapiens | A0A2R8Y7N0 | EPB41 | Unknown |
| 337 | H. sapiens | A0A087WWT8 | GDAP1L1 | Unknown |
| 338 | H. sapiens | K7EJ34 | RETREG3 | Unknown |
| 339 | H. sapiens | A0A0A0MRG8 | BAK1 | Unknown |
| 340 | H. sapiens | I3L376 | TVP23B | Unknown |
| 341 | H. sapiens | F5GYX3 | SEMA7A | Unknown |
| 342 | H. sapiens | Q5VTX9 | IFNLR1 | Unknown |
| 343 | H. sapiens | J3KP61 | PLD5 | Unknown |
| 344 | H. sapiens | A0A087X0T8 | CADM1 | Unknown |
| 345 | H. sapiens | J3KSN8 | SMIM21 | Unknown |
| 346 | H. sapiens | F8WAW2 | KIAA0319L | Unknown |
| 347 | H. sapiens | F8W1Z3 | CERS5 | Unknown |
| 348 | H. sapiens | A0A1B0GW78 | RASGEF1B | Unknown |
| 349 | H. sapiens | H7C593 | MFSD1 | Unknown |
| 350 | H. sapiens | A0A0G2JM16 | MUC4 | Unknown |
| 351 | H. sapiens | E9PM16 | ZNF7 | Unknown |
| 352 | H. sapiens | E5RFT6 | LYPLA1 | Unknown |
| 353 | H. sapiens | E9PM70 | CYB561D1 | Unknown |
| 354 | H. sapiens | C9JQU6 | ARL6IP5 | Unknown |
| 355 | H. sapiens | J3QLU8 | PEMT | Unknown |
| 356 | H. sapiens | E9PQQ2 | MYB | Unknown |
| 357 | H. sapiens | A0A0A0MRG3 | ZNF138 | Unknown |
| 358 | H. sapiens | A0A0D9SF04 | CLN3 | Unknown |
| 359 | H. sapiens | F8W782 | ADIPOR1 | Unknown |
| 360 | H. sapiens | F8WEP4 | CHL1 | Unknown |
| 361 | H. sapiens | J3KRT1 | DHX38 | Unknown |
| 362 | H. sapiens | B5MEG5 | USP19 | Unknown |
| 363 | H. sapiens | D6RHV8 | TMEM175 | Unknown |
| 364 | H. sapiens | I3L1G0 | SLC5A11 | Unknown |
| 365 | H. sapiens | F8WCS3 | POLR1B | Unknown |
| 366 | H. sapiens | A0A140TA65 | CES5A | Unknown |
| 367 | H. sapiens | F8WCU3 | SLC30A6 | Unknown |
| 368 | H. sapiens | D6R9B4 | CD164 | Unknown |
| 369 | H. sapiens | F8VXV4 | SLC48A1 | Unknown |
| 370 | H. sapiens | E9PIV8 | CKLF-CMTM1 | Unknown |
| 371 | H. sapiens | F8VWE0 | TSPAN31 | Unknown |
| 372 | H. sapiens | A0A1B0GUE0 | JAKMIP1 | Unknown |
| 373 | H. sapiens | F5H7K7 | LPCAT3 | Unknown |
| 374 | H. sapiens | D6RB93 | ZNF451 | Unknown |
| 375 | H. sapiens | E9PM26 | MS4A7 | Unknown |
| 376 | H. sapiens | F8WF83 | SLC9A9 | Unknown |
| 377 | H. sapiens | F5H7G2 | RGMA | Unknown |
| 378 | H. sapiens | E5RI04 | ANKRD46 | Unknown |
| 379 | H. sapiens | D6RJC0 | SLC41A3 | Unknown |
| 380 | H. sapiens | E9PJF1 | MYB | Unknown |
| 381 | H. sapiens | F8WEW7 | PORCN | Unknown |
| 382 | H. sapiens | F8WF33 | ARL6IP5 | Unknown |
| 383 | H. sapiens | J3KPT4 | TRABD | Unknown |
| 384 | H. sapiens | H0YL57 | RPLP1 | Unknown |
| 385 | H. sapiens | E9PMW8 | COP1 | Unknown |
| 386 | H. sapiens | A0A1B0GU12 | ATP6AP2 | Unknown |
| 387 | H. sapiens | E9PKL6 | OR51E1 | Unknown |
| 388 | H. sapiens | K7EPN3 | RAMP2 | Unknown |
| 389 | H. sapiens | A0A0G2JQ71 | ZNF66 | Unknown |
| 390 | H. sapiens | A0A0A0MTQ3 | CFAP54 | Unknown |
| 391 | H. sapiens | A0A0A0MSB7 | CALN1 | Unknown |
| 392 | H. sapiens | F8WDW0 | LMBR1 | Unknown |
| 393 | H. sapiens | A0A0U1RQZ5 | ENTPD1 | Unknown |
| 394 | H. sapiens | D6RI03 | TSPAN17 | Unknown |
| 395 | H. sapiens | V9GYR6 | ADPRM | Unknown |
| 396 | H. sapiens | J3QKR4 | ICAM2 | Unknown |
| 397 | H. sapiens | D6RC55 | OCIAD1 | Unknown |
| 398 | H. sapiens | C9JE17 | CCDC136 | Unknown |
| 399 | H. sapiens | C9JU31 | CCDC136 | Unknown |
| 400 | H. sapiens | F6VI00 | ACOT2 | Unknown |
| 401 | H. sapiens | H3BTX6 | ARL6IP1 | Unknown |
| 402 | H. sapiens | F8WCI3 | CDK5RAP2 | Unknown |
| 403 | H. sapiens | F8WB21 | SYS1 | Unknown |
| 404 | H. sapiens | A6NG31 | RARRES3 | Unknown |
| 405 | H. sapiens | F8WCL9 | ECE2 | Unknown |
| 406 | H. sapiens | A0A1B0GU51 | C14orf132 | Unknown |
| 407 | H. sapiens | J3KTR2 | PEMT | Unknown |
| 408 | H. sapiens | F8WDI1 | C3orf33 | Unknown |
| 409 | H. sapiens | F8WDN0 | URGCP | Unknown |
| 410 | H. sapiens | E9PN09 | SLC36A4 | Unknown |
| 411 | H. sapiens | F8VQZ6 | CERS5 | Unknown |
| 412 | H. sapiens | F8W0W6 | SNRPF | Unknown |
| 413 | H. sapiens | F5GX39 | TMED2 | Unknown |
| 414 | H. sapiens | F8WEN8 | LMBR1 | Unknown |
| 415 | H. sapiens | A0A0G2JN91 | NCR1 | Unknown |
| 416 | H. sapiens | F8WAW3 | GPR156 | Unknown |
| 417 | H. sapiens | D6RDM3 | SLC41A3 | Unknown |
| 418 | H. sapiens | D6RBY2 | TMEM33 | Unknown |
| 419 | H. sapiens | A0A0A6YYJ0 | MSANTD3-TMEFF1 | Unknown |
| 420 | H. sapiens | D6RBP2 | GYPB | Unknown |
| 421 | H. sapiens | A0A0G2JMZ5 | UGT2B15 | Unknown |
| 422 | H. sapiens | A0A075B785 | RELCH | Unknown |
| 423 | H. sapiens | A0A087WZR4 | FCGR3B | Unknown |
| 424 | H. sapiens | F8VSK7 | CERS5 | Unknown |
| 425 | H. sapiens | F5H0T7 | SLC22A6 | Unknown |
| 426 | H. sapiens | B5MC89 | THADA | Unknown |
| 427 | H. sapiens | E9PRZ6 | CDC27 | Unknown |
| 428 | H. sapiens | F5H5G1 | LSAMP | Unknown |
| 429 | H. sapiens | F8W1K4 | CERS5 | Unknown |
| 430 | H. sapiens | F6WFR7 | NTM | Unknown |
| 431 | H. sapiens | H3BP21 | NFAT5 | Unknown |
| 432 | H. sapiens | A0A2R8YF92 | SEL1L2 | Unknown |
| 433 | H. sapiens | E9PR36 | MTNR1B | Unknown |
| 434 | H. sapiens | F8WB98 | GGCX | Unknown |
| 435 | H. sapiens | C9JAX8 | SMIM4 | Unknown |
| 436 | H. sapiens | H3BS23 | MOSMO | Unknown |
| 437 | H. sapiens | A0A0D9SFD8 | CCDC163 | Unknown |
| 438 | H. sapiens | E9PFA2 | WDR17 | Unknown |
| 439 | H. sapiens | E7EQN9 | INPP4B | Unknown |
| 440 | H. sapiens | A0A087WX97 | BCL2L13 | Unknown |
| 441 | H. sapiens | E9PHR9 | PLSCR4 | Unknown |
| 442 | H. sapiens | F8WE64 | ELP6 | Unknown |
| 443 | H. sapiens | X6RLY7 | CACNA2D4 | Unknown |
| 444 | H. sapiens | F8WCA0 | VAMP2 | Unknown |
| 445 | H. sapiens | G3V5F3 | SCFD1 | Unknown |
| 446 | H. sapiens | H3BQA3 | PDPK1 | Unknown |
| 447 | H. sapiens | E9PM87 | PTPN22 | Unknown |
| 448 | H. sapiens | F8WDI5 | STIMATE | Unknown |
| 449 | H. sapiens | F5H4H7 | CLEC12B | Unknown |
| 450 | H. sapiens | K7ENK9 | VAMP2 | Unknown |
| 451 | H. sapiens | A0A087WT28 | CD200R1L | Unknown |
| 452 | H. sapiens | G3V232 | ADSSL1 | Unknown |
| 453 | H. sapiens | F8W1N7 | CERS5 | Unknown |
| 454 | H. sapiens | E9PRZ2 | PGAP2 | Unknown |
| 455 | H. sapiens | A0A182DWE8 | CFAP47 RP13-11B7.1 hCG_1982542 | Unknown |
| 456 | H. sapiens | Q8TDQ4 | TMEM222 | Unknown |
| 457 | H. sapiens | I3L1D2 | MPDU1 | Unknown |
| 458 | H. sapiens | A0A0G2JJ55 | MICA | Unknown |
| 459 | H. sapiens | E9PC20 | RAMP1 | Unknown |
| 460 | H. sapiens | H0YNL7 | PIGH | Unknown |
| 461 | H. sapiens | E9PKT4 | TMEM123 | Unknown |
| 462 | H. sapiens | G8JLJ3 | SMIM29 | Unknown |
| 463 | H. sapiens | G3V1A8 | LY6G6C hCG_43718 | Unknown |
| 464 | H. sapiens | A6NGS0 | UBE2J2 hCG_20420 | Unknown |
| 465 | H. sapiens | F5H3M3 | MANSC1 | Unknown |
| 466 | H. sapiens | K4JQN1 | BAX | Unknown |
| 467 | H. sapiens | A0A075B778 | ABCA5 | Unknown |
| 468 | H. sapiens | A0A1W2PR24 | ST3GAL5 | Unknown |
| 469 | H. sapiens | A0A0G2JP96 | LILRA1 | Unknown |
| 470 | H. sapiens | M9MML0 | FCGR3A | Unknown |
| 471 | H. sapiens | A0A2R8Y694 | SLC19A3 | Unknown |
| 472 | H. sapiens | F8WDB3 | ARF4 | Unknown |
| 473 | H. sapiens | F8WE00 | MFSD9 | Unknown |
| 474 | H. sapiens | J3QS78 | CD7 | Unknown |
| 475 | H. sapiens | D6RCD9 | TMEM175 | Unknown |
| 476 | H. sapiens | F2Z397 | TMEM184B | Unknown |
| 477 | H. sapiens | M0R1X3 | CEACAM8 | Unknown |
| 478 | H. sapiens | S4R453 | KCNMA1 | Unknown |
| 479 | H. sapiens | Q0P6N6 | NRG4 | Unknown |
| 480 | H. sapiens | F2Z2J3 | COA1 | Unknown |
| 481 | H. sapiens | I3L1Z6 | ABCC6 | Unknown |
| 482 | H. sapiens | F8WCB8 | FTO | Unknown |
| 483 | H. sapiens | K7ENB6 | SLC7A10 | Unknown |
| 484 | H. sapiens | F5H326 | LDHC | Unknown |
| 485 | H. sapiens | E9PKZ1 | SLC16A4 | Unknown |
| 486 | H. sapiens | M0R2F1 | KCNN4 | Unknown |
| 487 | H. sapiens | G3V5W3 | SOS2 | Unknown |
| 488 | H. sapiens | Q4KN23 | KIR3DS1 | Unknown |
| 489 | H. sapiens | G3V1I3 | FAM9C hCG_19162 | Unknown |
| 490 | H. sapiens | F8VRN7 | TMEM116 | Unknown |
| 491 | H. sapiens | A0A0G2JJ84 | BTNL2 | Unknown |
| 492 | H. sapiens | Q8WZ67 | KLRK1 | Unknown |
| 493 | H. sapiens | F5GWC9 | TMEM91 | Unknown |
| 494 | H. sapiens | B7Z596 | TPM1 | Unknown |
| 495 | H. sapiens | C9JKN6 | THSD7B | Unknown |
| 496 | H. sapiens | G3V248 | IFI27L1 | Unknown |
| 497 | H. sapiens | G3XAK3 | CLIP4 RSNL2 hCG_1783765 | Unknown |
| 498 | H. sapiens | A1A4Z5 | TRPC7 | Unknown |
| 499 | H. sapiens | C9J7K9 | PLSCR1 hCG_17108 | Unknown |
| 500 | H. sapiens | H3BUX2 | CYB5B | Unknown |
| 501 | H. sapiens | S4R3Y8 | TMEM91 | Unknown |
| 502 | H. sapiens | F5H5K1 | LRRC37B | Unknown |
| 503 | H. sapiens | A0A286YFJ5 | MFSD8 | Unknown |
| 504 | H. sapiens | F8W7G1 | CD200 | Unknown |
| 505 | H. sapiens | F8VVR0 | MRPL42 | Unknown |
| 506 | H. sapiens | A0A087X1Q6 | TARM1 | Unknown |
| 507 | H. sapiens | F8WCC4 | C3orf18 | Unknown |
| 508 | H. sapiens | K7EQ13 | G6PC3 | Unknown |
| 509 | H. sapiens | F8WEV1 | MAATS1 | Unknown |
| 510 | H. sapiens | A0A0A0MT53 | CD200R1L | Unknown |
| 511 | H. sapiens | E9PHY6 | LRRC8C | Unknown |
| 512 | H. sapiens | H7BXH0 | KCTD20 | Unknown |
| 513 | H. sapiens | E9PIJ2 | CYB561D1 | Unknown |
| 514 | H. sapiens | V9GYC5 | COMMD7 | Unknown |
| 515 | H. sapiens | B5MCI6 | MEMO1 | Unknown |
| 516 | H. sapiens | E9PQX4 | TDRKH | Unknown |
| 517 | H. sapiens | K7ELD9 | SYNGR2 | Unknown |
| 518 | H. sapiens | F5H038 | CLEC1A | Unknown |
| 519 | H. sapiens | K7EIN4 | TMED1 | Unknown |
| 520 | H. sapiens | Q5SNW4 | CLCN6 | Unknown |
| 521 | H. sapiens | E9PQJ6 | BET1L | Unknown |
| 522 | H. sapiens | F2Z2P5 | ERGIC1 | Unknown |
| 523 | H. sapiens | F8WDT4 | SUN3 | Unknown |
| 524 | H. sapiens | D6RE04 | PLRG1 | Unknown |
| 525 | H. sapiens | J3KST8 | CRLF3 | Unknown |
| 526 | H. sapiens | J3KRW3 | CEP95 | Unknown |
| 527 | H. sapiens | H3BNZ7 | C16orf95 | Unknown |
| 528 | H. sapiens | A2A2E0 | MANBAL | Unknown |
| 529 | H. sapiens | E7ETC6 | PDPN | Unknown |
| 530 | H. sapiens | A0A0H2UH41 | POTEM | Unknown |
| 531 | H. sapiens | I3L380 | ABHD12 | Unknown |
| 532 | H. sapiens | H0Y870 | TMEM222 | Unknown |
| 533 | H. sapiens | F8WCD4 | TMEM184B | Unknown |
| 534 | H. sapiens | A0A0A0MS18 | RAD51B | Unknown |
| 535 | H. sapiens | E5RK16 | FAXDC2 | Unknown |
| 536 | H. sapiens | A0A087WXA9 | KIZ | Unknown |
| 537 | H. sapiens | I3L072 | C17orf80 | Unknown |
| 538 | H. sapiens | K7EPU5 | SPRED3 | Unknown |
| 539 | H. sapiens | F8W1G5 | RNASEK | Unknown |
| 540 | H. sapiens | Q5T4Q8 | CD72 | Unknown |
| 541 | H. sapiens | A0A1W2PRT0 | ST3GAL5 | Unknown |
| 542 | H. sapiens | H3BU94 | SNAP23 | Unknown |
| 543 | H. sapiens | A0A2R8YEW2 | CYSTM1 ORF1-FL49 hCG_45310 | Unknown |
| 544 | H. sapiens | B3KT51 | TM2D3 hCG_26600 | Unknown |
| 545 | H. sapiens | E9PI46 | ABCD4 | Unknown |
| 546 | H. sapiens | B7Z863 | SLMAP | Unknown |
| 547 | H. sapiens | F8WEN7 | MTFP1 | Unknown |
| 548 | H. sapiens | A0A0C4DFN5 | TCTN3 C10orf61 hCG_39491 | Unknown |
| 549 | H. sapiens | M0QZX7 | ZNF816 | Unknown |
| 550 | H. sapiens | Q8N329 | EOGT C3orf64 | Unknown |
| 551 | H. sapiens | F2Z2A2 | MFSD9 | Unknown |
| 552 | H. sapiens | A0A1W2PQE2 | HLA-B | Unknown |
| 553 | H. sapiens | E5RG25 | UBE2W | Unknown |
| 554 | H. sapiens | A0A0J9YWY1 | LLCFC1 C7orf34 hCG_20688 | Unknown |
| 555 | H. sapiens | M0R0R3 | SMIM7 | Unknown |
| 556 | H. sapiens | A0AVG3 | TSNARE1 | Unknown |
| 557 | H. sapiens | B1ANB7 | MCOLN3 hCG_1775160 | Unknown |
| 558 | H. sapiens | I3L288 | TMEM159 LOC57146 hCG_38247 | Unknown |
| 559 | H. sapiens | B7Z964 | SLMAP | Unknown |
| 560 | H. sapiens | X6R3D1 | HRASLS hCG_16315 | Unknown |
| 561 | H. sapiens | E7EM61 | SLC19A3 | Unknown |
| 562 | H. sapiens | Q3KQS6 | MME | Unknown |
| 563 | H. sapiens | B9TX75 | MED24 | Unknown |
| 564 | H. sapiens | G5E972 | TMPO hCG_2015322 | Unknown |
| 565 | H. sapiens | F8VV56 | CD63 | Unknown |
| 566 | H. sapiens | D6RCL9 | SERF1B SERF1A | Unknown |
| 567 | H. sapiens | B3KT28 | FAF1 | Unknown |
| 568 | H. sapiens | G3V1R8 | TMBIM4 hCG_16706 | Unknown |
| 569 | H. sapiens | G5E9Q6 | PFN2 hCG_21343 | Unknown |
| 570 | H. sapiens | A0A0C4DGX8 | ATP6AP1 hCG_2008012 | Unknown |
| 571 | H. sapiens | K7EMW4 | NCLN hCG_23630 | Unknown |
| 572 | H. sapiens | B4DKD2 | ADAM11 | Unknown |
| 573 | H. sapiens | E5RGC5 | TVP23C-CDRT4 TVP23C | Unknown |
| 574 | S. cerevisiae | Q03941 | CAB5 YDR196C YD9346.07C | Both |
| 575 | S. cerevisiae | Q08215 | PEX15 PAS21 YOL044W | Both |
| 576 | S. cerevisiae | P25580 | PBN1 YCL052C YCL52C | ER |
| 577 | S. cerevisiae | P32854 | PEP12 VPS6 VPT13 YOR036W OR26.29 | ER |
| 578 | S. cerevisiae | Q05637 | PHM6 YDR281C D9954.14 | ER |
| 579 | S. cerevisiae | Q08931 | PRM3 YPL192C | ER |
| 580 | S. cerevisiae | P39926 | SSO2 YMR183C YM8010.13C | ER |
| 581 | S. cerevisiae | P31377 | SYN8 UIP2 YAL014C FUN34 | ER |
| 582 | S. cerevisiae | P32867 | SSO1 YPL232W P1405 | ER |
| 583 | S. cerevisiae | P31109 | SNC1 YAL030W | ER |
| 584 | S. cerevisiae | P33328 | SNC2 YOR327C | ER |
| 585 | S. cerevisiae | P38247 | SLM4 EGO3 GSE1 YBR077C YBR0723 | ER |
| 586 | S. cerevisiae | P43682 | SFT1 YKL006C-A YKL006BC | ER |
| 587 | S. cerevisiae | Q03322 | TLG1 YDR468C D8035.11 | ER |
| 588 | S. cerevisiae | P52870 | SBH1 SEB1 YER087C-B YER087BC | ER |
| 589 | S. cerevisiae | Q6Q595 | SCS22 YBL091C-A | ER |
| 590 | S. cerevisiae | P35179 | SSS1 YDR086C D4475 | ER |
| 591 | S. cerevisiae | P40075 | SCS2 YER120W | ER |
| 592 | S. cerevisiae | P22214 | SEC22 SLY2 TSL26 YLR268W L8479.3 | ER |
| 593 | S. cerevisiae | P52871 | SBH2 SEB2 YER019C-A YER019BC | ER |
| 594 | S. cerevisiae | Q01590 | SED5 YLR026C | ER |
| 595 | S. cerevisiae | P38342 | TSC10 YBR265W YBR1734 | ER |
| 596 | S. cerevisiae | Q12255 | NYV1 MAM2 YLR093C | ER |
| 597 | S. cerevisiae | P14020 | DPM1 SED3 YPR183W P9705.3 | ER |
| 598 | S. cerevisiae | Q08955 | CSM4 YPL200W | ER |
| 599 | S. cerevisiae | Q06001 | FAR10 YLR238W | ER |
| 600 | S. cerevisiae | P22804 | BET1 SLY12 YIL004C YIA4C | ER |
| 601 | S. cerevisiae | P25385 | BOS1 YLR078C L9449.9 | ER |
| 602 | S. cerevisiae | P40312 | CYB5 YNL111C N1949 | ER |
| 603 | S. cerevisiae | P38736 | GOS1 YHL031C | ER |
| 604 | S. cerevisiae | P32363 | SPT14 CWH6 GPI3 YPL175W P2269 | ER |
| 605 | S. cerevisiae | P43560 | LAM5 LTC2 YFL042C | ER |
| 606 | S. cerevisiae | P48353 | HLJ1 YMR161W YM8520.10 | ER |
| 607 | S. cerevisiae | Q99332 | FRT1 HPH1 YOR324C O6159 | ER |
| 608 | S. cerevisiae | P32339 | HMX1 YLR205C L8167.18 | ER |
| 609 | S. cerevisiae | Q3E790 | TSC3 YBR058C-A | ER |
| 610 | S. cerevisiae | Q04338 | VTI1 YMR197C YM9646.10C | ER |
| 611 | S. cerevisiae | Q3E842 | YMR122W-A | ER |
| 612 | S. cerevisiae | P38216 | YBR016W YBR0222 | ER |
| 613 | S. cerevisiae | P53146 | USE1 SLT1 YGL098W | ER |
| 614 | S. cerevisiae | P38374 | YSY6 YBR162W-A YBR162BW | ER |
| 615 | S. cerevisiae | Q05899 | YLR297W | ER |
| 616 | S. cerevisiae | Q03944 | VPS64 FAR9 YDR200C YD9346.10C | ER |
| 617 | S. cerevisiae | P33296 | UBC6 DOA2 YER100W | ER |
| 618 | S. cerevisiae | P41834 | UFE1 YOR075W YOR29-26 | ER |
| 619 | S. cerevisiae | Q08959 | PGC1 YPL206C | Mit |
| 620 | S. cerevisiae | P80967 | TOM5 MOM8A YPR133W-A | Mit |
| 621 | S. cerevisiae | P53507 | TOM7 MOM7 YNL070W N2378 | Mit |
| 622 | S. cerevisiae | P33448 | TOM6 ISP6 YOR045W | Mit |
| 623 | S. cerevisiae | P40515 | FIS1 MDV2 YIL065C | Mit |
| 624 | S. cerevisiae | P39722 | GEM1 YAL048C | Mit |
| 625 | S. cerevisiae | P22289 | QCR9 UCR9 YGR183C | MitIn |
| 626 | S. cerevisiae | P07255 | COX9 YDL067C D2520 | MitIn |
| 627 | S. cerevisiae | P10174 | COX7 YMR256C YM9920.10C | MitIn |
| 628 | S. cerevisiae | Q2V2P9 | YDR119W-A | MitIn |
| 629 | S. cerevisiae | Q02969 | PEX25 YPL112C | Pex |
| 630 | S. cerevisiae | P38335 | MTC4 YBR255W YBR1723 | Pex |
| 631 | S. cerevisiae | Q02820 | NCE101 NCE1 YJL205C YJL205BC YJL205C-A | Unknown |
| 632 | S. cerevisiae | Q03441 | RMD1 YDL001W | Unknown |
| 633 | S. cerevisiae | P43620 | RMD8 YFR048W | Unknown |
| 634 | S. cerevisiae | Q08559 | FYV12 YOR183W | Unknown |
| 635 | S. cerevisiae | P11927 | KAR1 YNL188W N1611 | Unknown |
| 636 | S. cerevisiae | Q08630 | IRC13 YOR235W | Unknown |
| 637 | S. cerevisiae | P0CD97 | YER039C-A | Unknown |
| 638 | S. cerevisiae | Q3E828 | YJL127C-B | Unknown |
| 639 | S. cerevisiae | Q8TGS8 | YMR105W-A | Unknown |
| 640 | S. cerevisiae | Q3E760 | YMR030W-A | Unknown |
| 641 | S. cerevisiae | O13511 | YAL065C | Unknown |
| 642 | S. cerevisiae | P39563 | YAR064W | Unknown |
| 643 | S. cerevisiae | Q2V2Q3 | YBR201C-A | Unknown |
| 644 | S. cerevisiae | Q3E743 | YJR112W-A | Unknown |
| 645 | S. cerevisiae | P47080 | YJL007C J1379 | Unknown |
| 646 | S. cerevisiae | P36092 | YKL044W YKL257 | Unknown |
| 647 | S. cerevisiae | Q2V2P2 | YKL065W-A | Unknown |
| 648 | S. cerevisiae | Q07738 | YDL241W | Unknown |
| 649 | S. cerevisiae | Q05612 | YDR278C | Unknown |
| 650 | S. cerevisiae | Q03480 | YDR209C YD8142A.06c | Unknown |
| 651 | S. cerevisiae | Q3E750 | YGL041C-B | Unknown |
| 652 | S. cerevisiae | Q8TGK1 | YHR213W-B | Unknown |
| 653 | S. cerevisiae | Q07074 | YHR007C-A | Unknown |
| 654 | S. cerevisiae | A5Z2X5 | YPR010C-A | Unknown |
| 655 | S. cerevisiae | P53229 | YGR045C | Unknown |
| 656 | S. cerevisiae | Q2V2P3 | YKL023C-A | Unknown |
| 657 | S. cerevisiae | Q3E814 | YLL006W-A | Unknown |
| 658 | S. cerevisiae | Q12506 | YOR314W 06123 | Unknown |
| 659 | S. cerevisiae | Q8TGU7 | YBR126W-A | Unknown |
| 660 | S. cerevisiae | Q04597 | YDR114C | Unknown |
| 661 | S. cerevisiae | P0C268 | YBL039W-B | Unknown |
| 662 | S. cerevisiae | Q96VH3 | YCL021W-A | Unknown |
| 663 | S. cerevisiae | Q8TGT9 | YGR146C-A | Unknown |
| 664 | S. cerevisiae | Q08110 | YOL014W | Unknown |
| 665 | S. cerevisiae | Q08734 | YOR268C | Unknown |
| 666 | S. cerevisiae | P53156 | YGL081W | Unknown |
| 667 | S. cerevisiae | Q05898 | YLR296W L8003.6 | Unknown |
| 668 | S. cerevisiae | P38185 | YBL071C YBL0615 | Unknown |
| i | Metric | Scale | AUROC | Best Threshold | TP | FP | TN | FN | Correct | Incorrect |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Patch 15 | Kyte & Doolittle | 96.28 | 29.40 | 38 | 0 | 10 | 5 | 48 | 5 |
| 2 | Patch 15 | TM Tendency | 95.81 | 13.42 | 36 | 0 | 10 | 7 | 46 | 7 |
| 3 | Patch 11 | Kyte & Doolittle | 94.88 | 25.50 | 35 | 0 | 10 | 8 | 45 | 8 |
| 4 | Wheel Face 5 | TM Tendency | 94.65 | 7.97 | 38 | 1 | 9 | 5 | 47 | 6 |
| 5 | Wheel Face 9 | TM Tendency | 93.95 | 12.31 | 37 | 0 | 10 | 6 | 47 | 6 |
| 6 | Patch 15 | GES | 93.95 | 5.09 | 32 | 0 | 10 | 11 | 42 | 11 |
| 7 | Patch 19 | Kyte & Doolittle | 93.49 | 28.30 | 34 | 0 | 10 | 9 | 44 | 9 |
| 8 | Wheel Face 7 | TM Tendency | 93.02 | 10.57 | 34 | 0 | 10 | 9 | 44 | 9 |
| 9 | Wheel Face 8 | TM Tendency | 92.91 | 11.04 | 36 | 0 | 10 | 7 | 46 | 7 |
| 10 | Segment 15 | Kyte & Doolittle | 92.56 | 35.20 | 31 | 0 | 10 | 12 | 41 | 12 |
| 11 | Segment 15 | TM Tendency | 92.33 | 16.28 | 33 | 0 | 10 | 10 | 43 | 10 |
| 12 | Patch 19 | TM Tendency | 91.86 | 13.32 | 35 | 0 | 10 | 8 | 45 | 8 |
| 13 | Segment 15 | GES | 91.63 | 5.43 | 35 | 0 | 10 | 8 | 45 | 8 |
| 14 | Segment 11 | Kyte & Doolittle | 91.63 | 27.10 | 33 | 0 | 10 | 10 | 43 | 10 |
| 15 | Rectangle 9 | Kyte & Doolittle | 91.28 | 18.10 | 31 | 0 | 10 | 12 | 41 | 12 |
| 16 | Wheel Face 5 | Kyte & Doolittle | 91.05 | 18.10 | 35 | 1 | 9 | 8 | 44 | 9 |
| 17 | Wheel Face 7 | Kyte & Doolittle | 91.05 | 21.00 | 38 | 2 | 8 | 5 | 46 | 7 |
| 18 | Rectangle 9 | Fauchere Pliska | 90.93 | 7.55 | 35 | 0 | 10 | 8 | 45 | 8 |
| 19 | Patch 19 | Octanol | 90.93 | 6.86 | 34 | 0 | 10 | 9 | 44 | 9 |
| 20 | Rectangle 9 | TM Tendency | 90.93 | 8.32 | 33 | 0 | 10 | 10 | 43 | 10 |
| 21 | Patch 19 | GES | 90.70 | 4.60 | 33 | 0 | 10 | 10 | 43 | 10 |
| 22 | Line 13 | Kyte & Doolittle | 90.70 | 13.90 | 37 | 2 | 8 | 6 | 45 | 8 |
| 23 | Patch 11 | GES | 90.35 | 4.32 | 31 | 0 | 10 | 12 | 41 | 12 |
| 24 | Star 8 | Kyte & Doolittle | 90.12 | 14.60 | 33 | 1 | 9 | 10 | 42 | 11 |
| 25 | TMD (18 aa) | Kyte & Doolittle | 90.00 | 42.90 | 27 | 0 | 10 | 16 | 37 | 16 |
| 26 | TMD | Kyte & Doolittle | 90.00 | 42.90 | 27 | 0 | 10 | 16 | 37 | 16 |
| 27 | Wheel Face 9 | GES | 89.88 | 4.46 | 32 | 0 | 10 | 11 | 42 | 11 |
| 28 | Patch 19 | Fauchere Pliska | 89.77 | 14.67 | 36 | 1 | 9 | 7 | 45 | 8 |
| 29 | Segment 19 | Kyte & Doolittle | 89.77 | 35.40 | 35 | 1 | 9 | 8 | 44 | 9 |
| 30 | Patch 11 | TM Tendency | 89.77 | 13.17 | 31 | 0 | 10 | 12 | 41 | 12 |
| 31 | Star 8 | TM Tendency | 89.77 | 6.94 | 35 | 1 | 9 | 8 | 44 | 9 |
| 32 | Wheel Face 8 | Kyte & Doolittle | 89.53 | 26.60 | 28 | 0 | 10 | 15 | 38 | 15 |
| 33 | Segment 19 | TM Tendency | 89.53 | 16.48 | 32 | 0 | 10 | 11 | 42 | 11 |
| 34 | Rectangle 9 | GES | 89.30 | 2.86 | 33 | 0 | 10 | 10 | 43 | 10 |
| 35 | Wheel Face 8 | GES | 89.19 | 4.10 | 31 | 0 | 10 | 12 | 41 | 12 |
| 36 | Line 13 | GES | 89.19 | 2.13 | 35 | 1 | 9 | 8 | 44 | 9 |
| 37 | Wheel Face 6 | TM Tendency | 89.07 | 9.79 | 32 | 0 | 10 | 11 | 42 | 11 |
| 38 | Segment 19 | Fauchere Pliska | 89.07 | 19.91 | 36 | 1 | 9 | 7 | 45 | 8 |
| 39 | Segment 11 | TM Tendency | 89.07 | 12.43 | 34 | 1 | 9 | 9 | 43 | 10 |
| 40 | TMD (18 aa) | TM Tendency | 88.84 | 16.45 | 34 | 0 | 10 | 9 | 44 | 9 |
| 41 | Line 13 | TM Tendency | 88.72 | 6.71 | 34 | 1 | 9 | 9 | 43 | 10 |
| 42 | Wheel Face 7 | GES | 88.37 | 3.87 | 27 | 0 | 10 | 16 | 37 | 16 |
| 43 | Wheel Face 9 | Roseman | 88.37 | 12.65 | 37 | 2 | 8 | 6 | 45 | 8 |
| 44 | Rectangle 9 | Roseman | 88.37 | 8.89 | 29 | 0 | 10 | 14 | 39 | 14 |
| 45 | TMD average | Kyte & Doolittle | 88.37 | 1.69 | 36 | 2 | 8 | 7 | 44 | 9 |
| 46 | Star 8 | Roseman | 88.26 | 6.75 | 36 | 2 | 8 | 7 | 44 | 9 |
| 47 | Wheel Face 9 | Kyte & Doolittle | 88.14 | 27.20 | 35 | 2 | 8 | 8 | 43 | 10 |
| 48 | Patch 15 | Fauchere Pliska | 88.14 | 15.50 | 31 | 0 | 10 | 12 | 41 | 12 |
| 49 | Patch 19 | Roseman | 88.14 | 15.30 | 31 | 0 | 10 | 12 | 41 | 12 |
| 50 | Patch 15 | Octanol | 88.14 | 7.89 | 32 | 0 | 10 | 11 | 42 | 11 |
| 51 | Wheel Face 7 | Roseman | 88.02 | 10.75 | 35 | 1 | 9 | 8 | 44 | 9 |
| 52 | Wheel Face 8 | Roseman | 87.91 | 12.57 | 30 | 0 | 10 | 13 | 40 | 13 |
| 53 | Star 8 | Fauchere Pliska | 87.91 | 6.97 | 25 | 0 | 10 | 18 | 35 | 18 |
| 54 | TMD | TM Tendency | 87.67 | 16.80 | 33 | 0 | 10 | 10 | 43 | 10 |
| 55 | Segment 11 | GES | 87.44 | 4.60 | 30 | 0 | 10 | 13 | 40 | 13 |
| 56 | Segment 15 | Fauchere Pliska | 87.44 | 17.09 | 32 | 1 | 9 | 11 | 41 | 12 |
| 57 | Star 8 | GES | 87.21 | 2.24 | 35 | 1 | 9 | 8 | 44 | 9 |
| 58 | Wheel Face 5 | GES | 87.09 | 2.64 | 38 | 3 | 7 | 5 | 45 | 8 |
| 59 | Wheel Face 4 | Kyte & Doolittle | 87.09 | 14.40 | 38 | 3 | 7 | 5 | 45 | 8 |
| 60 | Rectangle 9 | Octanol | 86.63 | 5.73 | 29 | 0 | 10 | 14 | 39 | 14 |
| 61 | Wheel Face 9 | Fauchere Pliska | 86.51 | 12.24 | 37 | 2 | 8 | 6 | 45 | 8 |
| 62 | Wheel Face 9 | Octanol | 86.28 | 7.10 | 36 | 2 | 8 | 7 | 44 | 9 |
| 63 | Segment 19 | Octanol | 86.28 | 9.65 | 32 | 0 | 10 | 11 | 42 | 11 |
| 64 | TMD average | TM Tendency | 86.28 | 0.88 | 31 | 0 | 10 | 12 | 41 | 12 |
| 65 | Wheel Face 6 | Kyte & Doolittle | 85.93 | 19.60 | 33 | 1 | 9 | 10 | 42 | 11 |
| 66 | TMD (18 aa) | GES | 85.93 | 5.78 | 34 | 1 | 9 | 9 | 43 | 10 |
| 67 | Patch 11 | Fauchere Pliska | 85.93 | 13.22 | 27 | 0 | 10 | 16 | 37 | 16 |
| 68 | Segment 11 | Fauchere Pliska | 85.81 | 13.72 | 31 | 1 | 9 | 12 | 40 | 13 |
| 69 | Star 8 | Octanol | 85.81 | 4.54 | 34 | 1 | 9 | 9 | 43 | 10 |
| 70 | TMD | GES | 85.81 | 5.78 | 34 | 1 | 9 | 9 | 43 | 10 |
| 71 | Twist 8 | Kyte & Doolittle | 85.70 | 18.50 | 35 | 2 | 8 | 8 | 43 | 10 |
| 72 | Patch 15 | Roseman | 85.35 | 15.46 | 31 | 1 | 9 | 12 | 40 | 13 |
| 73 | Segment 15 | Octanol | 85.35 | 8.78 | 32 | 1 | 9 | 11 | 41 | 12 |
| 74 | Wheel Face 8 | Octanol | 85.23 | 7.44 | 31 | 1 | 9 | 12 | 40 | 13 |
| 75 | Wheel Face 6 | GES | 84.88 | 3.39 | 28 | 1 | 9 | 15 | 37 | 16 |
| 76 | Segment 19 | GES | 84.65 | 6.61 | 27 | 0 | 10 | 16 | 37 | 16 |
| 77 | Twist 8 | Fauchere Pliska | 84.65 | 8.74 | 32 | 1 | 9 | 11 | 41 | 12 |
| 78 | Patch 11 | Roseman | 84.65 | 13.51 | 28 | 0 | 10 | 15 | 38 | 15 |
| 79 | Line 9 | Kyte & Doolittle | 84.30 | 11.10 | 36 | 2 | 8 | 7 | 44 | 9 |
| 80 | Twist 8 | TM Tendency | 84.19 | 9.59 | 29 | 0 | 10 | 14 | 39 | 14 |
| 81 | Wheel Face 8 | Fauchere Pliska | 84.07 | 10.93 | 35 | 2 | 8 | 8 | 43 | 10 |
| 82 | Twist 8 | GES | 83.95 | 2.93 | 35 | 2 | 8 | 8 | 43 | 10 |
| 83 | Segment 15 | Roseman | 83.95 | 18.21 | 27 | 0 | 10 | 16 | 37 | 16 |
| 84 | TMD average | Fauchere Pliska | 83.95 | 1.02 | 34 | 1 | 9 | 9 | 43 | 10 |
| 85 | TMD | Fauchere Pliska | 83.95 | 23.74 | 28 | 0 | 10 | 15 | 38 | 15 |
| 86 | Line 13 | Roseman | 83.72 | 6.76 | 33 | 1 | 9 | 10 | 42 | 11 |
| 87 | TMD average | GES | 83.72 | 0.29 | 33 | 1 | 9 | 10 | 42 | 11 |
| 88 | TMD (18 aa) | Fauchere Pliska | 83.49 | 23.74 | 27 | 0 | 10 | 16 | 37 | 16 |
| 89 | Wheel Face 7 | Fauchere Pliska | 83.26 | 10.17 | 35 | 2 | 8 | 8 | 43 | 10 |
| 90 | Segment 11 | Roseman | 83.02 | 12.94 | 33 | 2 | 8 | 10 | 41 | 12 |
| 91 | TMD (18 aa) | Octanol | 83.02 | 9.25 | 31 | 0 | 10 | 12 | 41 | 12 |
| 92 | TMD | Octanol | 83.02 | 9.25 | 31 | 0 | 10 | 12 | 41 | 12 |
| 93 | Wheel Face 7 | Octanol | 82.79 | 7.29 | 26 | 0 | 10 | 17 | 36 | 17 |
| 94 | Line 17 | Kyte & Doolittle | 82.79 | 14.60 | 30 | 1 | 9 | 13 | 39 | 14 |
| 95 | Patch 11 | Octanol | 82.56 | 7.67 | 32 | 1 | 9 | 11 | 41 | 12 |
| 96 | TMD average | Octanol | 82.56 | 0.41 | 32 | 1 | 9 | 11 | 41 | 12 |
| 97 | Segment 19 | Roseman | 82.21 | 19.04 | 29 | 1 | 9 | 14 | 38 | 15 |
| 98 | Wheel Face 5 | Fauchere Pliska | 81.40 | 7.57 | 37 | 3 | 7 | 6 | 44 | 9 |
| 99 | Wheel Face 6 | Roseman | 81.28 | 10.33 | 29 | 1 | 9 | 14 | 38 | 15 |
| 100 | Wheel Face 5 | Octanol | 80.93 | 5.84 | 27 | 1 | 9 | 16 | 36 | 17 |
| 101 | Wheel Face 5 | Roseman | 80.70 | 8.68 | 32 | 2 | 8 | 11 | 40 | 13 |
| 102 | Twist 8 | Roseman | 80.70 | 10.05 | 30 | 1 | 9 | 13 | 39 | 14 |
| 103 | Segment 11 | Octanol | 80.70 | 6.58 | 35 | 3 | 7 | 8 | 42 | 11 |
| 104 | Line 17 | TM Tendency | 80.70 | 7.06 | 30 | 1 | 9 | 13 | 39 | 14 |
| 105 | Wheel Face 6 | Fauchere Pliska | 80.47 | 8.73 | 37 | 2 | 8 | 6 | 45 | 8 |
| 106 | Wheel Face 4 | Octanol | 80.35 | 4.74 | 33 | 2 | 8 | 10 | 41 | 12 |
| 107 | TMD average | Roseman | 79.88 | 1.06 | 27 | 0 | 10 | 16 | 37 | 16 |
| 108 | TMD | Roseman | 79.88 | 22.76 | 25 | 0 | 10 | 18 | 35 | 18 |
| 109 | TMD (18 aa) | Roseman | 79.65 | 22.76 | 25 | 0 | 10 | 18 | 35 | 18 |
| 110 | Wheel Face 6 | Octanol | 79.42 | 6.25 | 32 | 2 | 8 | 11 | 40 | 13 |
| 111 | Wheel Face 4 | Fauchere Pliska | 79.30 | 6.50 | 37 | 3 | 7 | 6 | 44 | 9 |
| 112 | Line 17 | GES | 79.19 | 2.42 | 28 | 0 | 10 | 15 | 38 | 15 |
| 113 | Line 9 | GES | 78.60 | 1.79 | 34 | 3 | 7 | 9 | 41 | 12 |
| 114 | Twist 8 | Octanol | 76.86 | 6.32 | 28 | 1 | 9 | 15 | 37 | 16 |
| 115 | Wheel Face 3 | Kyte & Doolittle | 76.63 | 12.20 | 31 | 2 | 8 | 12 | 39 | 14 |
| 116 | Line 13 | Fauchere Pliska | 75.81 | 5.93 | 37 | 4 | 6 | 6 | 43 | 10 |
| 117 | Line 13 | Octanol | 74.88 | 4.08 | 31 | 3 | 7 | 12 | 38 | 15 |
| 118 | Wheel Face 4 | GES | 74.30 | 2.24 | 36 | 3 | 7 | 7 | 43 | 10 |
| 119 | Wheel Face 4 | TM Tendency | 74.07 | 6.65 | 40 | 3 | 7 | 3 | 47 | 6 |
| 120 | Line 9 | TM Tendency | 74.07 | 5.19 | 38 | 4 | 6 | 5 | 44 | 9 |
| 121 | Line 17 | Fauchere Pliska | 73.60 | 7.45 | 23 | 1 | 9 | 20 | 32 | 21 |
| 122 | Wheel Face 4 | Roseman | 72.44 | 7.21 | 36 | 4 | 6 | 7 | 42 | 11 |
| 123 | Wheel Face 3 | Fauchere Pliska | 70.58 | 5.27 | 32 | 3 | 7 | 11 | 39 | 14 |
| 124 | Line 17 | Octanol | 70.35 | 3.68 | 35 | 4 | 6 | 8 | 41 | 12 |
| 125 | Line 17 | Roseman | 68.37 | 7.39 | 27 | 3 | 7 | 16 | 34 | 19 |
| 126 | Wheel Face 3 | TM Tendency | 66.98 | 5.62 | 31 | 3 | 7 | 12 | 38 | 15 |
| 127 | Wheel Face 3 | GES | 66.05 | 1.80 | 36 | 5 | 5 | 7 | 41 | 12 |
| 128 | Wheel Face 3 | Roseman | 64.77 | 5.77 | 27 | 3 | 7 | 16 | 34 | 19 |
| 129 | Line 9 | Roseman | 64.19 | 5.87 | 19 | 1 | 9 | 24 | 28 | 25 |
| 130 | Wheel Face 3 | Octanol | 63.60 | 4.08 | 23 | 4 | 6 | 20 | 29 | 24 |
| 131 | Line 9 | Fauchere Pliska | 55.23 | 5.19 | 26 | 4 | 6 | 17 | 32 | 21 |
| 132 | TMD average | Roseman | 53.02 | 0.29 | 11 | 1 | 9 | 32 | 20 | 33 |
| 133 | TMD average | TM Tendency | 51.16 | 0.17 | 31 | 6 | 4 | 12 | 35 | 18 |
| 134 | TMD average | GES | -51.16 | 0.09 | 33 | 9 | 1 | 10 | 34 | 19 |
| 135 | TMD length | NA | -51.28 | 20.00 | 8 | 4 | 6 | 35 | 14 | 39 |
| 136 | Line 9 | Octanol | -51.40 | 4.08 | 22 | 7 | 3 | 21 | 25 | 28 |
| 137 | TMD average | Kyte & Doolittle | -53.26 | 0.68 | 34 | 10 | 0 | 9 | 34 | 19 |
| 138 | TMD average | Fauchere Pliska | -57.21 | 0.15 | 26 | 7 | 3 | 17 | 29 | 24 |
| 139 | TMD average | Octanol | -61.63 | 0.33 | 40 | 10 | 0 | 3 | 40 | 13 |
| Fig. 4 Ref # | Entry | TA Name | ER | Mito | Total # Of Cells | ER (%) | Mito (%) | Localization |
|---|---|---|---|---|---|---|---|---|
| 1 | P40515 | Fis1 | 0 | 33 | 33 | 0.00 | 1.00 | Mitochondria |
| 2 | Q2V2P9 | Cox26 | 2 | 18 | 20 | 0.10 | 0.90 | Mitochondria |
| 3 | Q2V2P3 | YKL023C | 131 | 3 | 134 | 0.98 | 0.02 | ER |
| 4 | P38185 | YBL071C | 122 | 3 | 125 | 0.98 | 0.02 | ER |
| 5 | Q05612 | YDR278C | 221 | 0 | 221 | 1.00 | 0.00 | ER |
| 6 | P0CD97 | YER039C | 161 | 0 | 161 | 1.00 | 0.00 | ER |
| 7 | P53156 | YGL081W | 135 | 0 | 135 | 1.00 | 0.00 | ER |
| 8 | Q12506 | YOR314W | 176 | 2 | 178 | 0.99 | 0.01 | ER |
| 9 | P53229 | YGR045C | 169 | 2 | 171 | 0.99 | 0.01 | ER |
| 10 | P36092 | YKL044W | 89 | 3 | 92 | 0.97 | 0.03 | ER |
| 11 | Q04597 | YDR114C | 1 | 141 | 142 | 0.01 | 0.99 | Mitochondria |
| 12 | Q03480 | YDR209C | 91 | 7 | 98 | 0.93 | 0.07 | ER |
| 13 | Q3E743 | YJR112W | 39 | 2 | 41 | 0.95 | 0.05 | ER |
| 14 | Q3E750 | YGL041C | 69 | 1 | 70 | 0.99 | 0.01 | ER |
| 15 | Q08110 | YOL014W | 91 | 7 | 98 | 0.93 | 0.07 | ER |
| 16 | Q05898 | YLR296W | 0 | 64 | 64 | 0.00 | 1.00 | Mitochondria |
| 17 | P0C268 | YBL039W | 38 | 8 | 46 | 0.83 | 0.17 | Other |
| i | Organism | Entry | Name | Localization |
|---|---|---|---|---|
| 1 | S. cerevisiae | Q08110 | YOL014W | ER |
| 2 | S. cerevisiae | P11927 | KAR1 | ER |
| 3 | S. cerevisiae | Q07738 | YDL241W | ER |
| 4 | S. cerevisiae | Q3E750 | YGL041C-B | ER |
| 5 | S. cerevisiae | Q8TGU7 | YBR126W-A | ER |
| 6 | S. cerevisiae | P43620 | RMD8 | ER |
| 7 | S. cerevisiae | Q07074 | YHR007C-A | ER |
| 8 | S. cerevisiae | Q08734 | YOR268C | ER |
| 9 | S. cerevisiae | Q3E743 | YJR112W-A | ER |
| 10 | S. cerevisiae | Q3E828 | YJL127C-B | Mit |
| 11 | S. cerevisiae | P36092 | YKL044W | Mit |
| 12 | S. cerevisiae | Q2V2P2 | YKL065W-A | Pex |
| 13 | S. cerevisiae | P0C268 | YBL039W-B | ER |
| 14 | S. cerevisiae | Q96VH3 | YCL021W-A | ER |
| 15 | S. cerevisiae | Q03441 | RMD1 | ER |
| i | Metric | Scale | AUROC | Best Threshold | TP | FP | TN | FN | Correct | Incorrect |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Wheel Face 7 | TM Tendency | 89.06 | 10.71 | 44 | 4 | 10 | 16 | 54 | 20 |
| 2 | Wheel Face 5 | TM Tendency | 87.64 | 8.38 | 45 | 1 | 13 | 20 | 58 | 21 |
| 3 | Patch 19 | Fauchere Pliska | 86.85 | 14.67 | 48 | 2 | 12 | 17 | 60 | 19 |
| 4 | Segment 11 | Kyte & Doolittle | 86.79 | 27.90 | 43 | 1 | 13 | 17 | 56 | 18 |
| 5 | Wheel Face 9 | TM Tendency | 86.51 | 12.68 | 43 | 1 | 13 | 21 | 56 | 22 |
| 6 | Wheel Face 6 | TM Tendency | 86.39 | 9.79 | 40 | 2 | 12 | 17 | 52 | 19 |
| 7 | Segment 15 | Kyte & Doolittle | 86.34 | 31.80 | 50 | 1 | 13 | 17 | 63 | 18 |
| 8 | Wheel Face 8 | TM Tendency | 86.34 | 12.16 | 40 | 0 | 14 | 29 | 54 | 29 |
| 9 | Patch 19 | Octanol | 86.05 | 7.02 | 44 | 2 | 12 | 20 | 56 | 22 |
| 10 | Wheel Face 5 | Kyte & Doolittle | 85.94 | 17.70 | 47 | 2 | 12 | 18 | 59 | 20 |
| 11 | Wheel Face 7 | Kyte & Doolittle | 85.94 | 21.20 | 51 | 3 | 11 | 15 | 62 | 18 |
| 12 | Patch 15 | Kyte & Doolittle | 85.71 | 31.00 | 46 | 3 | 11 | 12 | 57 | 15 |
| 13 | Wheel Face 9 | Fauchere Pliska | 85.60 | 12.78 | 44 | 0 | 14 | 25 | 58 | 25 |
| 14 | Patch 15 | Fauchere Pliska | 85.26 | 14.69 | 47 | 2 | 12 | 14 | 59 | 16 |
| 15 | Patch 15 | TM Tendency | 85.09 | 14.16 | 45 | 0 | 14 | 19 | 59 | 19 |
| 16 | Rectangle 9 | Fauchere Pliska | 84.86 | 7.96 | 46 | 1 | 13 | 19 | 59 | 20 |
| 17 | Patch 11 | Kyte & Doolittle | 84.75 | 25.50 | 46 | 1 | 13 | 15 | 59 | 16 |
| 18 | Segment 15 | TM Tendency | 84.47 | 16.28 | 41 | 1 | 13 | 18 | 54 | 19 |
| 19 | Patch 15 | Octanol | 84.13 | 7.78 | 45 | 1 | 13 | 23 | 58 | 24 |
| 20 | Segment 19 | Fauchere Pliska | 84.01 | 19.91 | 49 | 1 | 13 | 15 | 62 | 16 |
| 21 | Star 8 | Kyte & Doolittle | 84.01 | 14.60 | 48 | 2 | 12 | 26 | 60 | 28 |
| 22 | Wheel Face 5 | GES | 83.73 | 2.78 | 44 | 0 | 14 | 32 | 58 | 32 |
| 23 | Wheel Face 6 | Kyte & Doolittle | 83.67 | 19.40 | 48 | 3 | 11 | 15 | 59 | 18 |
| 24 | Rectangle 9 | TM Tendency | 83.50 | 8.23 | 42 | 0 | 14 | 20 | 56 | 20 |
| 25 | Star 8 | Fauchere Pliska | 83.50 | 6.99 | 30 | 0 | 14 | 26 | 44 | 26 |
| 26 | Wheel Face 7 | Fauchere Pliska | 83.45 | 10.17 | 49 | 3 | 11 | 16 | 60 | 19 |
| 27 | Wheel Face 7 | GES | 83.45 | 3.30 | 53 | 3 | 11 | 18 | 64 | 21 |
| 28 | Segment 11 | Fauchere Pliska | 83.11 | 13.97 | 39 | 0 | 14 | 29 | 53 | 29 |
| 29 | Rectangle 9 | Kyte & Doolittle | 82.99 | 18.10 | 40 | 4 | 10 | 15 | 50 | 19 |
| 30 | Segment 15 | Fauchere Pliska | 82.99 | 16.10 | 48 | 2 | 12 | 16 | 60 | 18 |
| 31 | Wheel Face 5 | Fauchere Pliska | 82.77 | 7.84 | 47 | 2 | 12 | 14 | 59 | 16 |
| 32 | Rectangle 9 | Octanol | 82.65 | 5.33 | 43 | 0 | 14 | 29 | 57 | 29 |
| 33 | Line 13 | GES | 82.65 | 2.30 | 40 | 6 | 8 | 15 | 48 | 21 |
| 34 | Patch 15 | GES | 82.65 | 5.09 | 38 | 4 | 10 | 10 | 48 | 14 |
| 35 | Rectangle 9 | Roseman | 82.60 | 8.43 | 43 | 3 | 11 | 14 | 54 | 17 |
| 36 | Wheel Face 7 | Octanol | 82.54 | 7.18 | 40 | 3 | 11 | 21 | 51 | 24 |
| 37 | Wheel Face 9 | Octanol | 82.54 | 8.82 | 34 | 0 | 14 | 23 | 48 | 23 |
| 38 | Wheel Face 8 | Fauchere Pliska | 82.48 | 11.57 | 46 | 1 | 13 | 17 | 59 | 18 |
| 39 | Patch 19 | Roseman | 82.43 | 13.34 | 43 | 1 | 13 | 25 | 56 | 26 |
| 40 | Wheel Face 7 | Roseman | 82.43 | 11.46 | 40 | 3 | 11 | 25 | 51 | 28 |
| 41 | Segment 11 | TM Tendency | 82.31 | 13.75 | 36 | 0 | 14 | 24 | 50 | 24 |
| 42 | Segment 11 | GES | 82.20 | 4.58 | 37 | 1 | 13 | 20 | 50 | 21 |
| 43 | Wheel Face 9 | Roseman | 82.20 | 12.65 | 49 | 1 | 13 | 27 | 62 | 28 |
| 44 | Wheel Face 6 | Fauchere Pliska | 82.14 | 9.00 | 48 | 0 | 14 | 33 | 62 | 33 |
| 45 | Segment 15 | GES | 81.97 | 5.43 | 43 | 0 | 14 | 25 | 57 | 25 |
| 46 | Line 13 | TM Tendency | 81.80 | 6.71 | 45 | 4 | 10 | 15 | 55 | 19 |
| 47 | Wheel Face 4 | Fauchere Pliska | 81.80 | 6.64 | 45 | 3 | 11 | 15 | 56 | 18 |
| 48 | Patch 11 | Fauchere Pliska | 81.75 | 13.22 | 34 | 2 | 12 | 15 | 46 | 17 |
| 49 | TMD average | Fauchere Pliska | 81.75 | 1.01 | 46 | 2 | 12 | 23 | 58 | 25 |
| 50 | Wheel Face 8 | Roseman | 81.63 | 12.67 | 38 | 0 | 14 | 31 | 52 | 31 |
| 51 | Wheel Face 8 | Octanol | 81.58 | 7.44 | 43 | 1 | 13 | 21 | 56 | 22 |
| 52 | Line 13 | Kyte & Doolittle | 81.41 | 14.90 | 40 | 2 | 12 | 38 | 52 | 40 |
| 53 | Patch 11 | TM Tendency | 81.41 | 13.17 | 39 | 0 | 14 | 23 | 53 | 23 |
| 54 | Rectangle 9 | GES | 81.35 | 2.86 | 40 | 0 | 14 | 24 | 54 | 24 |
| 55 | Wheel Face 9 | GES | 81.24 | 4.46 | 39 | 2 | 12 | 21 | 51 | 23 |
| 56 | Star 8 | TM Tendency | 81.18 | 6.92 | 46 | 1 | 13 | 20 | 59 | 21 |
| 57 | Wheel Face 6 | GES | 81.18 | 3.02 | 47 | 2 | 12 | 17 | 59 | 19 |
| 58 | Wheel Face 8 | Kyte & Doolittle | 81.18 | 22.80 | 53 | 2 | 12 | 26 | 65 | 28 |
| 59 | Star 8 | GES | 81.12 | 2.23 | 46 | 2 | 12 | 21 | 58 | 23 |
| 60 | Wheel Face 9 | Kyte & Doolittle | 81.12 | 24.80 | 48 | 2 | 12 | 26 | 60 | 28 |
| 61 | Wheel Face 8 | GES | 81.01 | 4.10 | 36 | 2 | 12 | 23 | 48 | 25 |
| 62 | Segment 19 | Kyte & Doolittle | 80.95 | 35.40 | 45 | 2 | 12 | 21 | 57 | 23 |
| 63 | Twist8 | Kyte & Doolittle | 80.95 | 18.50 | 48 | 0 | 14 | 23 | 62 | 23 |
| 64 | Twist8 | Fauchere Pliska | 80.90 | 8.74 | 47 | 0 | 14 | 17 | 61 | 17 |
| 65 | Patch 11 | GES | 80.78 | 4.03 | 43 | 3 | 11 | 16 | 54 | 19 |
| 66 | Patch 19 | TM Tendency | 80.61 | 13.32 | 44 | 0 | 14 | 23 | 58 | 23 |
| 67 | Segment 19 | TM Tendency | 80.50 | 16.48 | 43 | 1 | 13 | 19 | 56 | 20 |
| 68 | Segment 19 | Octanol | 80.33 | 9.65 | 43 | 0 | 14 | 19 | 57 | 19 |
| 69 | Star 8 | Roseman | 80.33 | 6.75 | 51 | 1 | 13 | 27 | 64 | 28 |
| 70 | Star 8 | Octanol | 80.27 | 4.17 | 52 | 0 | 14 | 23 | 66 | 23 |
| 71 | TMD | Fauchere Pliska | 80.16 | 22.95 | 38 | 4 | 10 | 25 | 48 | 29 |
| 72 | Wheel Face 4 | Octanol | 80.10 | 4.74 | 48 | 2 | 12 | 20 | 60 | 22 |
| 73 | TMD (18aa) | Fauchere Pliska | 79.93 | 22.95 | 37 | 5 | 9 | 15 | 46 | 20 |
| 74 | Line 13 | Roseman | 79.88 | 6.76 | 46 | 3 | 11 | 23 | 57 | 26 |
| 75 | Segment 15 | Octanol | 79.71 | 8.78 | 43 | 1 | 13 | 18 | 56 | 19 |
| 76 | Wheel Face 6 | Octanol | 79.31 | 6.25 | 43 | 3 | 11 | 11 | 54 | 14 |
| 77 | Patch 19 | Kyte & Doolittle | 79.20 | 28.30 | 42 | 5 | 9 | 10 | 51 | 15 |
| 78 | Patch 15 | Roseman | 79.14 | 15.16 | 41 | 1 | 13 | 23 | 54 | 24 |
| 79 | Wheel Face 4 | Kyte & Doolittle | 79.08 | 16.30 | 27 | 2 | 12 | 13 | 39 | 15 |
| 80 | Patch 11 | Octanol | 79.02 | 6.18 | 51 | 2 | 12 | 20 | 63 | 22 |
| 81 | Wheel Face 6 | Roseman | 78.63 | 10.33 | 38 | 4 | 10 | 12 | 48 | 16 |
| 82 | Segment 11 | Octanol | 78.57 | 9.52 | 30 | 4 | 10 | 12 | 40 | 16 |
| 83 | Patch 19 | GES | 78.46 | 4.92 | 33 | 0 | 14 | 27 | 47 | 27 |
| 84 | TMD | Kyte & Doolittle | 78.23 | 38.30 | 37 | 6 | 8 | 13 | 45 | 19 |
| 85 | TMD (18aa) | Kyte & Doolittle | 78.17 | 38.30 | 37 | 1 | 13 | 23 | 50 | 24 |
| 86 | Patch 11 | Roseman | 78.12 | 13.51 | 36 | 1 | 13 | 25 | 49 | 26 |
| 87 | TMD average | Kyte & Doolittle | 77.83 | 1.82 | 37 | 0 | 14 | 33 | 51 | 33 |
| 88 | Wheel Face 5 | Octanol | 77.83 | 5.79 | 37 | 1 | 13 | 20 | 50 | 21 |
| 89 | TMD average | Octanol | 77.55 | 0.40 | 42 | 5 | 9 | 13 | 51 | 18 |
| 90 | Segment 15 | Roseman | 77.32 | 18.21 | 35 | 2 | 12 | 22 | 47 | 24 |
| 91 | Line 9 | GES | 77.10 | 1.79 | 49 | 2 | 12 | 19 | 61 | 21 |
| 92 | Wheel Face 5 | Roseman | 76.87 | 9.03 | 30 | 1 | 13 | 30 | 43 | 31 |
| 93 | Line 17 | Fauchere Pliska | 76.76 | 7.26 | 40 | 2 | 12 | 18 | 52 | 20 |
| 94 | TMD | Octanol | 76.76 | 9.25 | 40 | 3 | 11 | 39 | 51 | 42 |
| 95 | TMD (18aa) | Octanol | 76.76 | 9.25 | 40 | 4 | 10 | 17 | 50 | 21 |
| 96 | Segment 11 | Roseman | 76.64 | 12.91 | 44 | 1 | 13 | 27 | 57 | 28 |
| 97 | TMD (18aa) | TM Tendency | 76.53 | 16.45 | 43 | 2 | 12 | 18 | 55 | 20 |
| 98 | Line 17 | TM Tendency | 76.13 | 7.06 | 40 | 5 | 9 | 8 | 49 | 13 |
| 99 | Segment 19 | GES | 75.85 | 6.61 | 31 | 0 | 14 | 30 | 45 | 30 |
| 100 | Line 17 | Kyte & Doolittle | 75.74 | 16.70 | 30 | 0 | 14 | 36 | 44 | 36 |
| 101 | TMD | TM Tendency | 75.74 | 16.80 | 42 | 3 | 11 | 21 | 53 | 24 |
| 102 | TMD average | TM Tendency | 75.74 | 0.94 | 34 | 2 | 12 | 23 | 46 | 25 |
| 103 | Wheel Face 4 | TM Tendency | 75.62 | 6.81 | 55 | 0 | 14 | 22 | 69 | 22 |
| 104 | Line 17 | GES | 75.57 | 2.49 | 32 | 4 | 10 | 11 | 42 | 15 |
| 105 | Line 9 | Kyte & Doolittle | 75.51 | 11.10 | 50 | 2 | 12 | 16 | 62 | 18 |
| 106 | Line 13 | Fauchere Pliska | 75.28 | 6.25 | 48 | 4 | 10 | 17 | 58 | 21 |
| 107 | Segment 19 | Roseman | 75.00 | 20.18 | x | 1 | 13 | 20 | 13 | 21 |
| 108 | Line 9 | TM Tendency | 74.72 | 5.40 | 42 | 1 | 13 | 18 | 55 | 19 |
| 109 | Twist8 | TM Tendency | 74.66 | 8.72 | 47 | 0 | 14 | 21 | 61 | 21 |
| 110 | Line 13 | Octanol | 74.60 | 4.08 | 46 | 5 | 9 | 30 | 55 | 35 |
| 111 | Twist8 | GES | 74.38 | 2.93 | 45 | 0 | 14 | 23 | 59 | 23 |
| 112 | Wheel Face 4 | Roseman | 74.26 | 7.38 | 37 | 1 | 13 | 28 | 50 | 29 |
| 113 | Wheel Face 3 | Fauchere Pliska | 74.21 | 5.20 | 46 | 1 | 13 | 24 | 59 | 25 |
| 114 | TMD average | Roseman | 74.09 | 1.06 | 32 | 7 | 7 | 11 | 39 | 18 |
| 115 | Wheel Face 4 | GES | 74.04 | 2.24 | 52 | 0 | 14 | 20 | 66 | 20 |
| 116 | TMD | Roseman | 73.64 | 19.92 | 36 | 1 | 13 | 34 | 49 | 35 |
| 117 | TMD (18aa) | Roseman | 73.53 | 19.92 | 36 | 2 | 12 | 17 | 48 | 19 |
| 118 | Line 17 | Octanol | 73.36 | 3.68 | 50 | 3 | 11 | 15 | 61 | 18 |
| 119 | TMD (18aa) | GES | 73.19 | 5.78 | 42 | 0 | 14 | 23 | 56 | 23 |
| 120 | TMD | GES | 73.02 | 5.78 | 42 | 4 | 10 | 14 | 52 | 18 |
| 121 | TMD average | GES | 72.56 | 0.29 | 40 | 0 | 14 | 31 | 54 | 31 |
| 122 | Wheel Face 3 | TM Tendency | 72.05 | 5.42 | 48 | 0 | 14 | 27 | 62 | 27 |
| 123 | Wheel Face 3 | Kyte & Doolittle | 70.52 | 12.50 | 25 | 0 | 14 | 20 | 39 | 20 |
| 124 | Twist8 | Octanol | 70.29 | 6.25 | 42 | 1 | 13 | 20 | 55 | 21 |
| 125 | Line 9 | Roseman | 70.07 | 5.87 | 29 | 0 | 14 | 33 | 43 | 33 |
| 126 | Wheel Face 3 | Roseman | 69.78 | 5.77 | 40 | 3 | 11 | 19 | 51 | 22 |
| 127 | Line 17 | Roseman | 69.50 | 6.14 | 52 | 2 | 12 | 26 | 64 | 28 |
| 128 | Twist8 | Roseman | 68.48 | 10.05 | 38 | 1 | 13 | 20 | 51 | 21 |
| 129 | Wheel Face 3 | GES | 66.89 | 1.80 | 48 | 0 | 14 | 26 | 62 | 26 |
| 130 | Wheel Face 3 | Octanol | 66.67 | 4.08 | 33 | 0 | 14 | 33 | 47 | 33 |
| 131 | Line 9 | Fauchere Pliska | 64.34 | 5.19 | 38 | 3 | 11 | 16 | 49 | 19 |
| 132 | Line 9 | Octanol | 59.24 | 4.08 | 24 | 2 | 12 | 26 | 36 | 28 |
| 133 | TMD len | NA | -53.23 | 20.00 | 11 | 5 | 9 | 52 | 20 | 57 |
| i | Metric | Scale | AUROC | Best Threshold | TP | FP | TN | FN | Correct | Incorrect |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Patch 11 | Kyte & Doolittle | 82.63 | 28.00 | 94 | 3 | 42 | 70 | 136 | 73 |
| 2 | Segment 19 | Kyte & Doolittle | 82.61 | 38.20 | 114 | 8 | 37 | 50 | 151 | 58 |
| 3 | Segment 11 | Kyte & Doolittle | 82.06 | 27.80 | 111 | 6 | 39 | 53 | 150 | 59 |
| 4 | Patch 11 | TM Tendency | 81.74 | 12.66 | 107 | 4 | 41 | 57 | 148 | 61 |
| 5 | Patch 15 | TM Tendency | 81.51 | 14.71 | 107 | 2 | 43 | 57 | 150 | 59 |
| 6 | Patch 15 | Kyte & Doolittle | 81.23 | 33.40 | 93 | 3 | 42 | 71 | 135 | 74 |
| 7 | Segment 11 | TM Tendency | 80.75 | 12.90 | 101 | 3 | 42 | 63 | 143 | 66 |
| 8 | TMD | Kyte & Doolittle | 80.72 | 32.10 | 142 | 15 | 30 | 22 | 172 | 37 |
| 9 | Patch 19 | TM Tendency | 80.59 | 13.13 | 125 | 9 | 36 | 39 | 161 | 48 |
| 10 | TMD (18 aa) | Kyte & Doolittle | 80.53 | 32.10 | 143 | 15 | 30 | 21 | 173 | 36 |
| 11 | Segment 15 | Kyte & Doolittle | 79.78 | 34.90 | 112 | 7 | 38 | 52 | 150 | 59 |
| 12 | Patch 19 | Kyte & Doolittle | 79.55 | 30.10 | 113 | 10 | 35 | 51 | 148 | 61 |
| 13 | Segment 15 | TM Tendency | 79.24 | 17.16 | 86 | 1 | 44 | 78 | 130 | 79 |
| 14 | TMD | TM Tendency | 79.17 | 18.86 | 106 | 9 | 36 | 58 | 142 | 67 |
| 15 | TMD (18 aa) | TM Tendency | 78.91 | 18.86 | 106 | 9 | 36 | 58 | 142 | 67 |
| 16 | Wheel Face 8 | Kyte & Doolittle | 78.90 | 22.70 | 131 | 16 | 29 | 33 | 160 | 49 |
| 17 | TMD (avg) | Kyte & Doolittle | 78.89 | 1.71 | 129 | 14 | 31 | 35 | 160 | 49 |
| 18 | Wheel Face 9 | Kyte & Doolittle | 78.88 | 26.60 | 113 | 11 | 34 | 51 | 147 | 62 |
| 19 | Segment 19 | TM Tendency | 78.40 | 19.68 | 85 | 1 | 44 | 79 | 129 | 80 |
| 20 | Wheel Face 9 | TM Tendency | 77.47 | 13.05 | 94 | 4 | 41 | 70 | 135 | 74 |
| 21 | Wheel Face 8 | TM Tendency | 77.23 | 11.32 | 116 | 13 | 32 | 48 | 148 | 61 |
| 22 | Wheel Face 7 | Kyte & Doolittle | 77.19 | 23.30 | 99 | 10 | 35 | 65 | 134 | 75 |
| 23 | Wheel Face 7 | TM Tendency | 76.99 | 11.10 | 88 | 5 | 40 | 76 | 128 | 81 |
| 24 | Rectangle 9 | TM Tendency | 76.76 | 7.98 | 110 | 9 | 36 | 54 | 146 | 63 |
| 25 | Wheel Face 6 | Kyte & Doolittle | 76.52 | 21.20 | 91 | 8 | 37 | 73 | 128 | 81 |
| 26 | Wheel Face 6 | TM Tendency | 76.44 | 9.28 | 109 | 10 | 35 | 55 | 144 | 65 |
| 27 | Twist 8 | Kyte & Doolittle | 75.75 | 20.90 | 95 | 5 | 40 | 69 | 135 | 74 |
| 28 | Star 8 | TM Tendency | 75.71 | 7.08 | 99 | 9 | 36 | 65 | 135 | 74 |
| 29 | Wheel Face 5 | Kyte & Doolittle | 75.55 | 19.20 | 82 | 4 | 41 | 82 | 123 | 86 |
| 30 | Twist 8 | TM Tendency | 75.37 | 9.58 | 89 | 5 | 40 | 75 | 129 | 80 |
| 31 | TMD (avg) | TM Tendency | 75.06 | 0.92 | 96 | 11 | 34 | 68 | 130 | 79 |
| 32 | Rectangle 9 | Kyte & Doolittle | 74.84 | 18.00 | 100 | 9 | 36 | 64 | 136 | 73 |
| 33 | Wheel Face 4 | Kyte & Doolittle | 72.90 | 15.60 | 88 | 11 | 34 | 76 | 122 | 87 |
| 34 | Line 9 | TM Tendency | 72.76 | 5.35 | 123 | 17 | 28 | 41 | 151 | 58 |
| 35 | Wheel Face 5 | TM Tendency | 70.75 | 9.10 | 71 | 5 | 40 | 93 | 111 | 98 |
| 36 | Wheel Face 4 | TM Tendency | 70.65 | 7.08 | 111 | 17 | 28 | 53 | 139 | 70 |
| 37 | Line 13 | Kyte & Doolittle | 70.51 | 12.70 | 130 | 19 | 26 | 34 | 156 | 53 |
| 38 | Line 9 | Kyte & Doolittle | 69.50 | 11.10 | 126 | 21 | 24 | 38 | 150 | 59 |
| 39 | Star 8 | Kyte & Doolittle | 69.32 | 14.90 | 106 | 15 | 30 | 58 | 136 | 73 |
| 40 | Line 17 | Kyte & Doolittle | 69.28 | 14.00 | 122 | 19 | 26 | 42 | 148 | 61 |
| 41 | Line 13 | TM Tendency | 69.00 | 7.18 | 78 | 7 | 38 | 86 | 116 | 93 |
| 42 | Line 17 | TM Tendency | 67.77 | 6.91 | 117 | 17 | 28 | 47 | 145 | 64 |
| 43 | Wheel Face 3 | TM Tendency | 65.30 | 5.40 | 134 | 25 | 20 | 30 | 154 | 55 |
| 44 | Wheel Face 3 | Kyte & Doolittle | 64.40 | 12.60 | 66 | 6 | 39 | 98 | 105 | 104 |
| 45 | TMD length | NA | 59.14 | 21.00 | 27 | 8 | 37 | 137 | 64 | 145 |
| 46 | CTE negative charge | NA | -61.06 | 11.00 | 164 | 45 | 0 | 0 | 164 | 45 |
| 47 | CTE net charge | NA | -62.47 | -2.00 | 3 | 3 | 42 | 161 | 45 | 164 |
| 48 | CTE positive charge | NA | -72.68 | 8.00 | 162 | 45 | 0 | 2 | 162 | 47 |
