5 Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis
Adapted from Barlow AN, Manu MS, Saladi SM, Tarr PT, Yadav Y, Thinn AMM, Zhu Y, Laganowsky AD, Clemons WM, Ramasamy S. 2023. Structures of Get3d reveal a distinct architecture associated with the emergence of photosynthesis. J Biol Chem 299:104752. doi:10.1016/j.jbc.2023.104752
5.1 Abstract
Homologs of the protein Get3 have been identified in all domains yet remain to be fully characterized. In the eukaryotic cytoplasm, Get3 delivers tail-anchored (TA) integral membrane proteins, defined by a single transmembrane helix at their C terminus, to the endoplasmic reticulum. While most eukaryotes have a single Get3 gene, plants are notable for having multiple Get3 paralogs. Get3d is conserved across land plants and photosynthetic bacteria and includes a distinctive C-terminal \(\alpha\)-crystallin domain. After tracing the evolutionary origin of Get3d, we solve the Arabidopsis thaliana Get3d crystal structure, identify its localization to the chloroplast, and provide evidence for a role in TA protein binding. The structure is identical to that of a cyanobacterial Get3 homolog, which is further refined here. Distinct features of Get3d include an incomplete active site, a “closed” conformation in the apo-state, and a hydrophobic chamber. Both homologs have ATPase activity and are capable of binding TA proteins, supporting a potential role in TA protein targeting. Get3d is first found with the development of photosynthesis and conserved across 1.2 billion years into the chloroplasts of higher plants across the evolution of photosynthesis suggesting a role in the homeostasis of photosynthetic machinery.
Keywords: cyanobacteria, GET pathway, Get3, protein targeting, protein structure, plant, NTPase, structural biology, tail-anchored protein
5.2 Introduction
A central problem for eukaryotes and photosynthetic microbes is the presence of multiple membranes that contain specific subsets of integral membrane proteins (IMPs). The correct targeting and insertion of IMPs to distinct and specific locations is necessary to maintain cellular homeostasis (Hegde and Keenan, 2022). Most IMPs span the lipid bilayer with hydrophobic \(\alpha\)-helical transmembrane domains (TMDs) that present a challenge during biogenesis, as they must be shielded from the aqueous environment prior to insertion to avoid aggregation. For IMPs, a signal, often the first TMD, encodes the destination, and the location of the signal in the protein sequence dictates cotranslational versus post-translational targeting.
A subset of IMPs, termed tail-anchored (TA) proteins, are defined by a single TMD within ~30 residues of the C terminus (Kutay et al., 1993), which serves as their targeting signal. Because of the position of their TMDs in the sequence, TA proteins must be fully synthesized and released by the ribosome prior to targeting (Shan, 2019). TA proteins are targeted in a post-translational chaperone-assisted mechanism primarily by the Guided Entry of TA proteins (GET) pathway. A central player in the GET pathway is Get3, which captures the TA signal in the cytosol and delivers the protein to a membrane insertion complex at the endoplasmic reticulum (ER), dependent on ATP hydrolysis (Chio et al., 2017; Fry et al., 2022; Rome et al., 2013). Get3 has been shown to have a conserved mechanism across eukaryotes (Fry et al., 2022; Rome et al., 2013). Many TA proteins play essential roles, for example, vesicle trafficking, protein localization, and regulation of apoptosis, and are targeted to various membranes (Abell and Mullen, 2011; Manu et al., 2018).
Organisms that generate energy via photosynthesis require additional cellular compartments and membranes, which necessitates added complexity for the targeting of TA proteins to these membranes (Frain et al., 2016). Examples are chloroplasts and other plastids, multimembrane organelles, derived from a single endosymbiotic event in which a cyanobacterial ancestor was incorporated into an early eukaryote, around 1.2 billion years ago (Oborník, 2019). Given this, it is unsurprising that protein targeting in chloroplasts has features conserved from protein targeting in bacteria. While much is known about membrane protein targeting in chloroplasts (Lee et al., 2017; Ries et al., 2020), it is unclear if a distinct mechanism exists for targeting of TA proteins to either the thylakoid or inner envelope membrane of chloroplasts. Interesting chloroplastic TA protein examples are the two SecE paralogs of the distinct Sec translocons present on either the inner envelope (SECE2) or thylakoid (SECE1) membranes where the respective targeting was found to be dependent solely on the TMD and flanking C-terminal residues (Anderson et al., 2019). Recently, the Get3 paralog AtGet3b has been implicated in targeting SECE1 to the thylakoid membrane (Anderson et al., 2021); however, further studies will be necessary to probe the role of AtGet3b in vivo. While the protein factors and specific targeting mechanisms are not yet known, sorting mechanisms are required that distinguish the characteristics of the TA proteins (Anderson et al., 2021; Fry et al., 2021).
Sequence analysis revealed that the Get3/ArsA fold family includes homologs from all three domains of life including one identified by Pfam in 2006 (El-Gebali et al., 2019) and discussed first by Chartron et al. in 2012 (Chartron et al., 2012) with a distinct architecture characterized by an \(\alpha\)-crystallin domain (\(\alpha\)CD) at its C terminus. Unlike fungi and metazoa, which contain a single Get3 in each genome, plants and cyanobacteria have multiple Get3 genes (Bodensohn et al., 2019; Chartron et al., 2012; Duncan et al., 2013; Manu et al., 2018; Xing et al., 2017). The model plant Arabidopsis thaliana (A. thaliana) contains four such genes (noted Get3a, b, c, and d). Evidence supports that AtGet3a (UniProt: Q949M9) resides in the cytosol, AtGet3b (UniProt: A1L4Y1) in the chloroplast stroma, and AtGet3c (UniProt: Q5XF80) in the mitochondrial matrix (Berardini et al., 2015; Bodensohn et al., 2019; Duncan et al., 2013; UniProt Consortium, 2021; Xing et al., 2017). AtGet3a has the same function as other cytoplasmic Get3 homologs in eukaryotes, targeting hydrophobic TA proteins in the cytosol to the ER membrane with knockouts of AtGet3a resulting in distinct phenotypes (Srivastava et al., 2017; Xing et al., 2017). There is conflicting evidence as to whether AtGet3b knockouts cause a phenotypic defect (Anderson et al., 2021; Xing et al., 2017), and no phenotype has been found for knockouts of AtGet3c (Xing et al., 2017). No experimental information is available for AtGet3d (UniProt: Q6DYE4).
Here, we provide a detailed characterization of Get3d, a distinct member of the Get3 family. We demonstrate that it first evolved in photosynthetic bacteria and has been conserved in the chloroplasts of plants. We solve the atomic structure of a plant Get3d and further refine a previously deposited structure of a cyanobacterial Get3d. We identify conserved functional motifs and identify distinct features of Get3d. We then investigate these functional motifs and show that Get3d can bind TA proteins irrespective of the unique \(\alpha\)CD at its C terminus and can hydrolyze ATP. This work provides a comprehensive characterization of a Get3 family member that has deep evolutionary roots connected to photosynthesis.
5.3 Results
5.3.1 Placing plant paralogs within the Get3/ArsA fold family
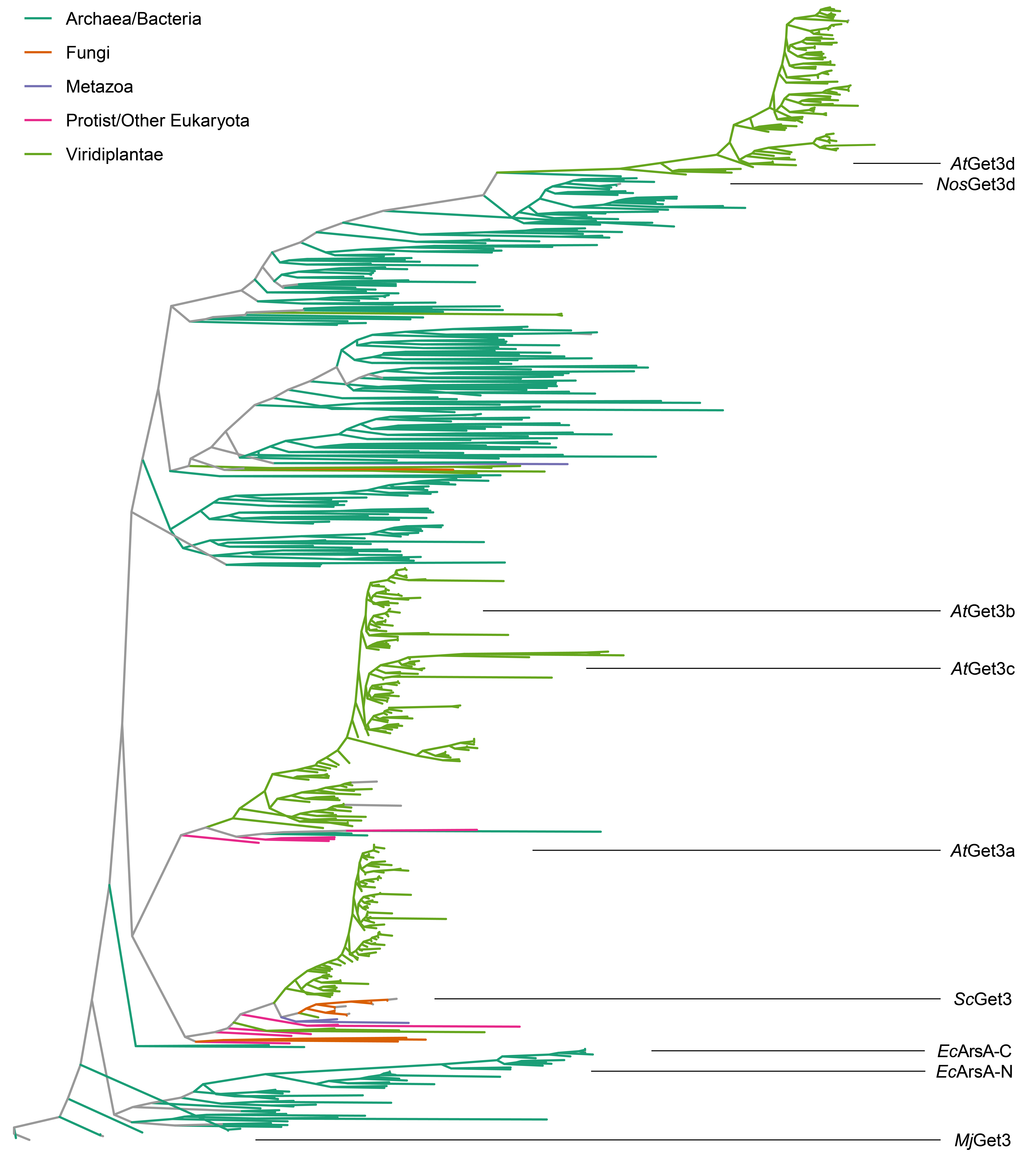
As Get3/ArsA homologs span the tree of life with several distinct clade lineages, we first sought to examine the evolutionary history of Get3d by performing a thorough phylogenetic reconstruction of Get3/ArsA homologs (Figure 5.1A and Supplementary Figure 5.1). We identified all Get3 proteins present in UniProt using the Pfam database (El-Gebali et al., 2019; UniProt Consortium, 2021) and then aligned them to a seed structural alignment of Get3 proteins based on solved 3D structures (Katoh and Standley, 2013; Russell and Barton, 1992). A phylogenetic reconstruction was then calculated using maximum likelihood with clades collapsed at a 70% bootstrap support (Stamatakis, 2014).
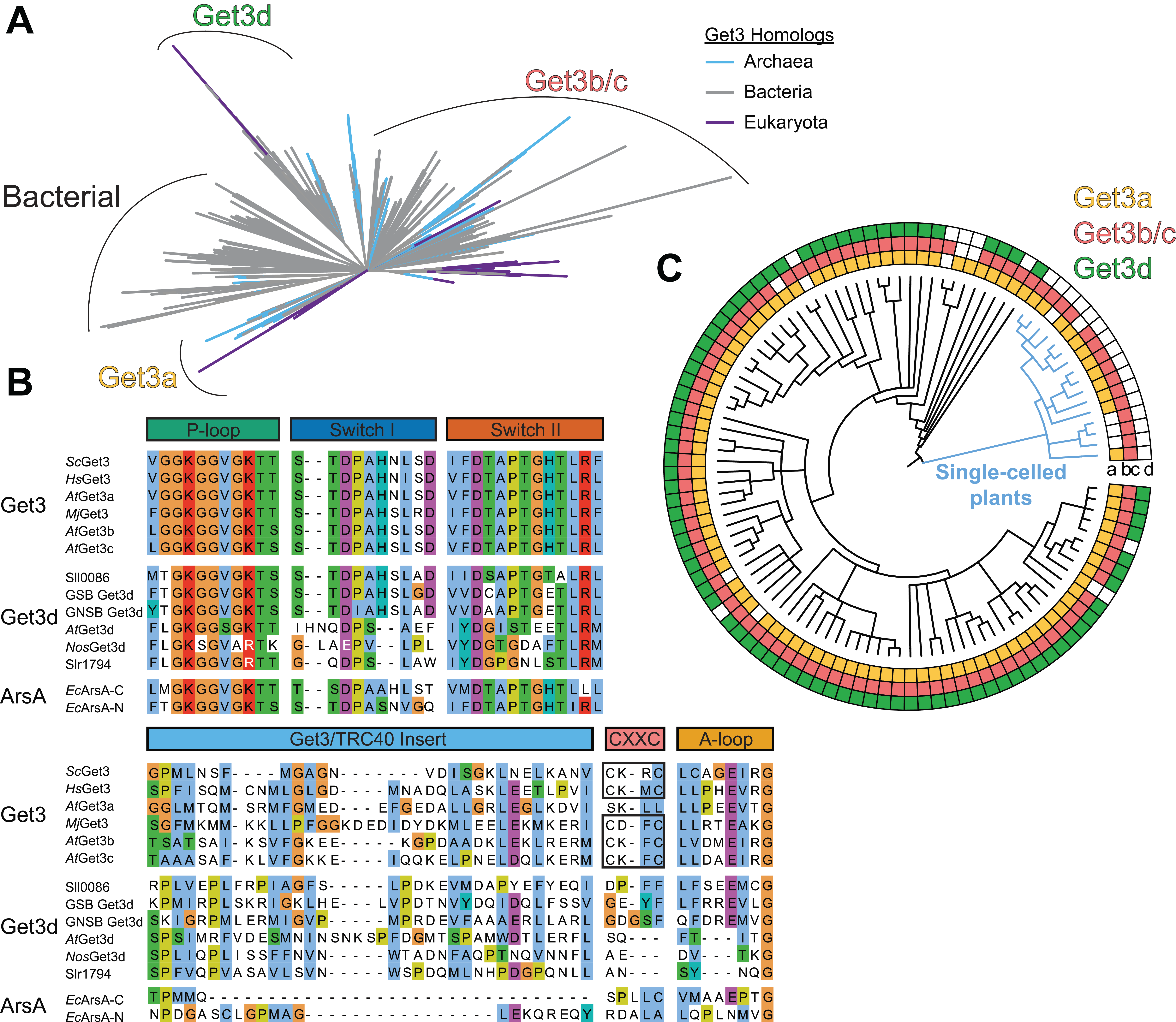
The first clade is ArsA, a soluble ATPase involved in protecting against arsenite toxicity found primarily in bacteria (Chen et al., 1986; Koonin, 1993; Leipe et al., 2002; Rosen et al., 1988; Zhou et al., 2000). Next is the well-studied cytoplasmic Get3 that is present across eukaryotes and some archaea and includes AtGet3a. Another Get3 lineage is restricted to Viridiplantae and contains both AtGet3b and AtGet3c. The final Get3 lineage is first found in green and purple bacteria and then cyanobacteria and across Viridiplantae and contains both AtGet3d and the Get3d homolog from the cyanobacteria Nostoc sp. PCC 7120 (NosGet3d) with a solved structure (Figure 5.1A and Supplementary Figure 5.1).
At the sequence level, Get3d proteins contain regions that can be aligned to notable motifs of the Get3 family, including the P-loop, switch-I, switch-II, Get3 motif/TRC40-insert, and A-loop (Figure 5.1B) (Suloway et al., 2009). The P-loop is well conserved with the so-called intradimeric (or “deviant”) Walker A motif (Koonin, 1993; Leipe et al., 2002; Maggiolo et al., 2023). Some Get3d proteins, such as the second homolog in Synechocystis sp. PCC 6803 (Sll0086) and green sulfur and nonsulfur Get3d, have well-conserved switch I, switch II, and A-loop regions. More common are homologs such as AtGet3d and NosGet3d that have degenerate catalytic residues including divergent switch I loops and missing A-loops. The hydrophobic nature of the Get3 motif/TRC40-insert of Get3 homologs is conserved across the Get3d family, unlike the related ArsA proteins (Suloway et al., 2009). The signature CXXC motif that coordinates a Zn2+ at the Get3 dimer interface is also missing in the Get3d family (Suloway et al., 2009).
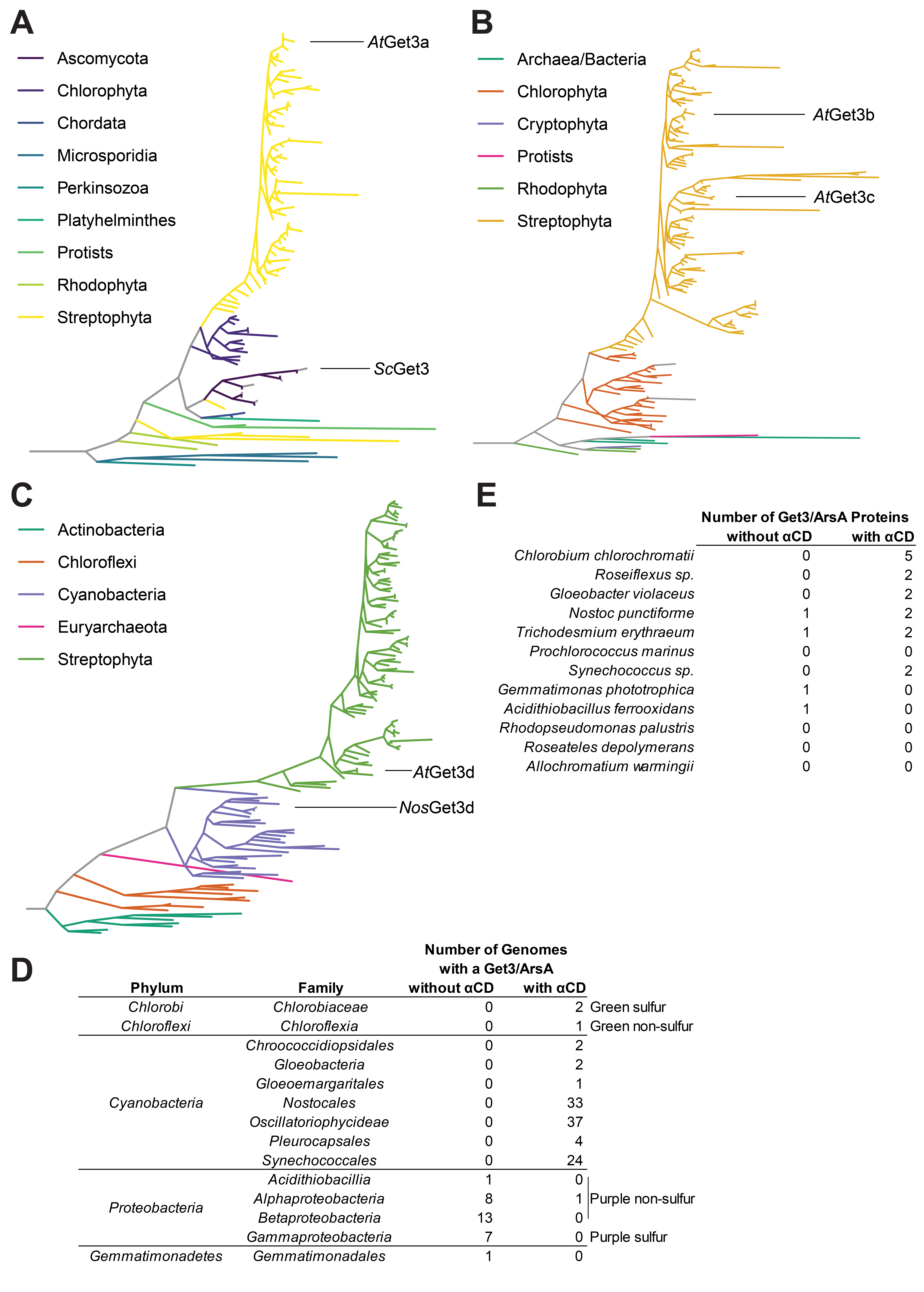
To better understand the origin and distribution of Get3 proteins from plants and photosynthetic bacteria, we carried out a more extensive phylogenetic reconstruction by specifically focusing on Get3 proteins from these two groups. Plant Get3a, Get3b/Get3c, as well as Get3d homologs, each form separate clades (Supplementary Figure 5.2A-C). Get3a forms a single clade with fungal and metazoan Get3 homologs found at their root (Supplementary Figure 5.1 and Supplementary Figure 5.2A). Get3b and Get3c trace to a single common ancestor, that is, monophyletic, and the clade cannot be split into separate b and c groups (Supplementary Figure 5.1 and Supplementary Figure 5.1B). Get3d forms a single clade with cyanobacteria and the green sulfur Chloroflexi Get3 proteins as the nearest relatives (Supplementary Figure 5.1 and Supplementary Figure 5.2C). Get3a and Get3b/c clades trace to a more recent common ancestor before their common ancestor with Get3d (Supplementary Figure 5.1).
To investigate the conservation of Get3d in photosynthetic bacteria specifically, we quantified the number of sequenced genomes that contain Get3 proteins with the characteristic \(\alpha\)CD in various phyla (Supplementary Figure 5.2D). Get3d is completely conserved in cyanobacteria and green sulfur and nonsulfur bacteria, which do not contain another Get3-like homolog. In purple bacteria, many families contain species that encode a cytoplasmic Get3 homolog, whereas only a single purple nonsulfur bacterium encodes a Get3d homolog. Furthermore, the number of Get3d homologs encoded by representative species from these phyla was determined (Supplementary Figure 5.2E). While some species contain no Get3d proteins, others contain multiple copies of Get3d in their genome, such as Chlorobium chlorochromatii, which contains five copies of Get3d. This could be due to whole genome duplication events (Serres et al., 2009) and horizontal gene transfers (Raymond and Blankenship, 2003; Zhaxybayeva et al., 2006), which are integral to the evolution of protein families and homologs (Shi and Falkowski, 2008). As the availability of sequenced genomes from photosynthetic bacteria increases, a broader depth of information about the evolution and conservation of Get3d in photosynthetic bacteria can be learned.
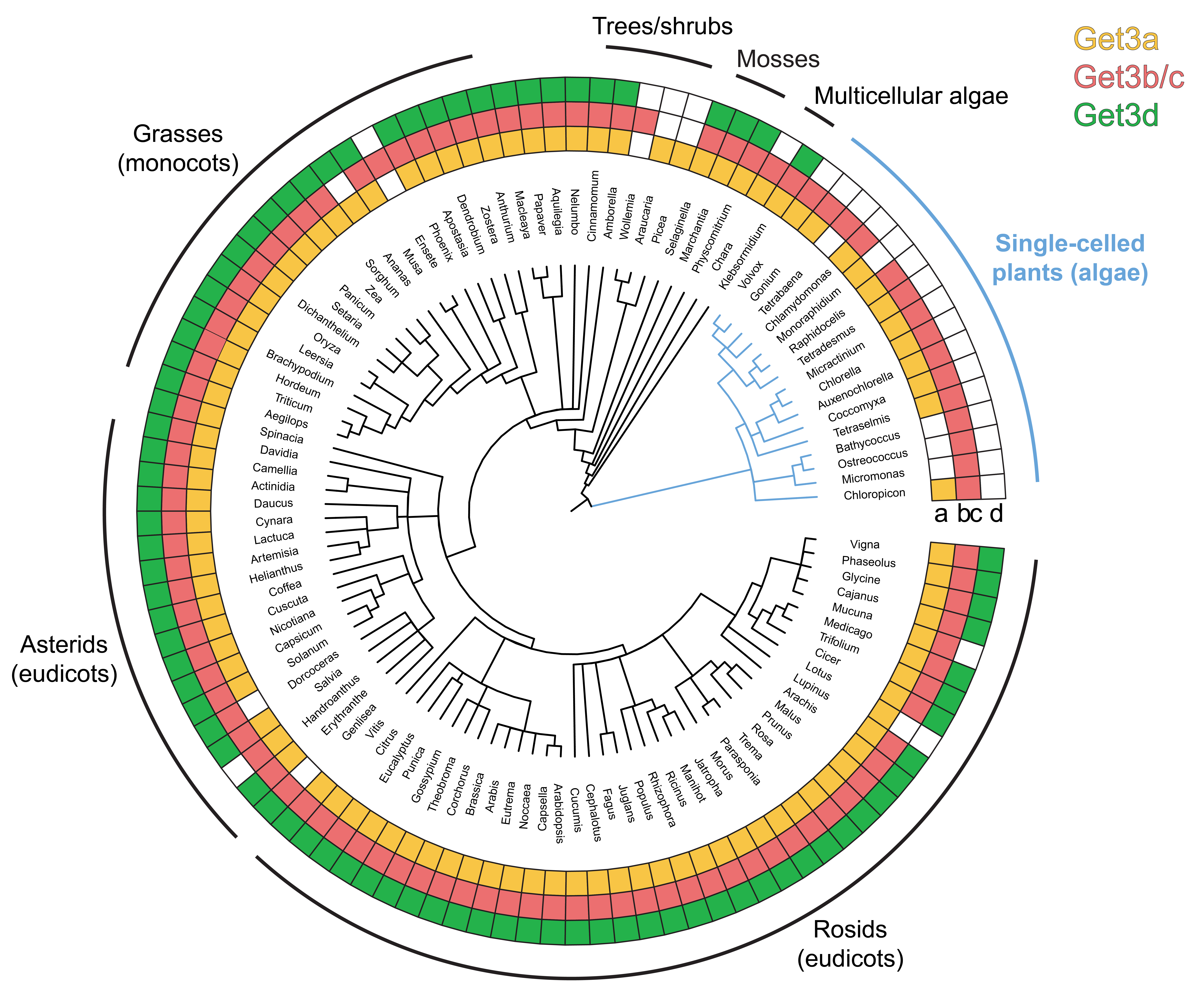
We next consider the distribution of the Get3 homologs across plants. Using our phylogenetic information, we assigned each Get3 to either the a, b/c, or d group, we can correlate the taxonomic distribution of each group across Viridiplantae (Figure 5.1C and Supplementary Figure 5.3). The tree is collapsed at the level of taxonomic genus to minimize errors resulting from uneven genome annotation; however, in some cases, poor annotation within a genus may preclude the ability to identify a given homolog. The results suggest that at least one protein from each of the Get3a and Get3b/c groups is present across all plant genomes. Get3d proteins are found across land plants (e.g., mosses, grasses, and eudicots) yet are completely missing in single-celled plants, the blue branch in Figure 5.1C and Supplementary Figure 5.3. Given that the nearest relative of Get3d is from a cyanobacteria and plants are derived from an endosymbiotic event that led to a chloroplast (Moore et al., 2019; Oborník, 2019; Zimorski et al., 2014), the absence of Get3d in single-celled plants suggests gene loss in the corresponding taxa.
5.3.2 Subcellular localization of Get3d
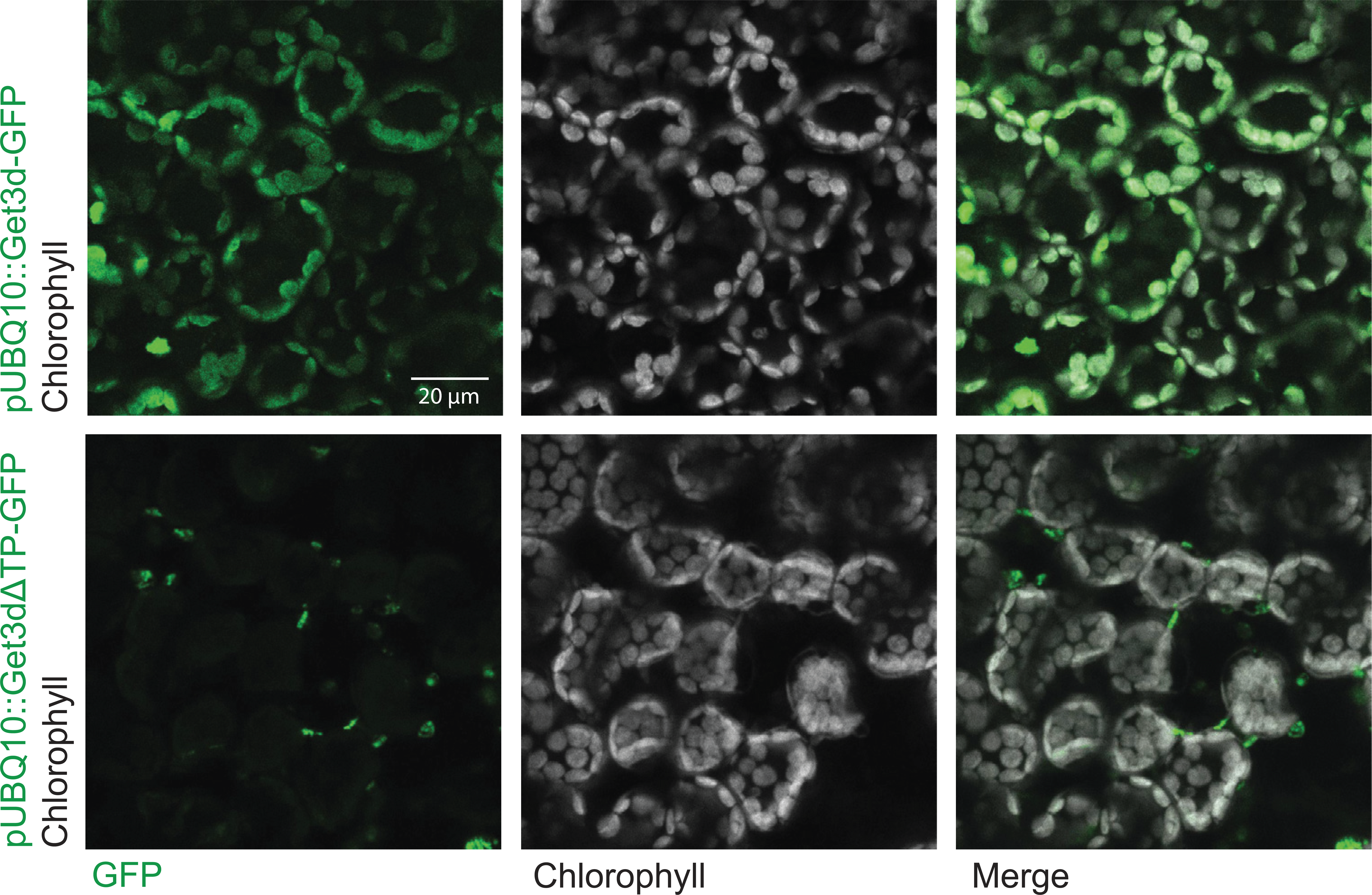
The cellular localization of AtGet3d has not been experimentally determined. Like many essential genes from the ancestral cyanobacteria, Get3d acquired a chloroplast targeting signal during the endosymbiotic transfer of genes into the plant genome (Moore et al., 2019; Oborník, 2019; Zimorski et al., 2014). Computational methods support this predicting that AtGet3d is localized to the chloroplast stroma (~91% likelihood) with a small probability (~5%) of it being localized to the thylakoid space (Almagro Armenteros et al., 2019).
To experimentally confirm the localization of AtGet3d, we employed Agrobacterium-mediated expression of AtGet3d in Nicotiana benthamiana leaves (Gelvin, 2017). Constructs were generated with GFP appended to the Get3d gene, with or without the predicted chloroplast transit peptide, as a C-terminal fusion. This allowed us to monitor the localization of expressed Get3d after Agrobacterium-mediated insertion into the Nicotiana genome. Upon infiltration, the tobacco leaves were monitored by fluorescence microscopy. For the full-length Get3d gene that contained the transit peptide, the fluorescence colocalized with the intrinsic chlorophyll autofluorescence, indicating that AtGet3d localizes to the chloroplast (Figure 5.2). Localization to the stroma versus thylakoid lumen could not be distinguished here. For Get3d lacking the transit peptide, the GFP signal gave a pattern of distinct puncta not associated with a clear subcellular structure in the mesophyll cells. The puncta are not consistent with the typical localization of GFP, the cytosol and nucleus, in these cells (Köhler, 1998). These results confirm the predictions of a functioning chloroplast targeting signal.
5.4 Discussion
In eukaryotes, the efficient and precise insertion of membrane proteins is an imperative step for their accurate function in various organelles (Fry and Clemons, 2018; Hegde and Keenan, 2022; Kunze and Berger, 2015). Errors in targeting may lead to mislocalization of these proteins, which can result in unfavorable cellular effects. Recent work explores the GET pathway in plants, with all components of the GET pathway excluding Get5 and Sgt2 having been identified (Asseck et al., 2021; Bodensohn et al., 2019; Duncan et al., 2013; Srivastava et al., 2017; Xing et al., 2017). However, a striking difference in plants compared with other eukaryotes is the presence of multiple paralogs of Get3 (Chartron et al., 2012; Duncan et al., 2013; Manu et al., 2018). In A. thaliana, four paralogs of Get3 exist, termed Get3a–d. This study is the first to characterize Get3d, a distinct homolog that is conserved across a few billion years of the evolution of photosynthesis from bacteria to plants.
Overall, some Get3d proteins are more similar in sequence to the cytoplasmic Get3 proteins, whereas others are more diverged (Figure 5.1B) suggesting distinct evolutionary paths. For example, Get3d proteins from representative green sulfur and green nonsulfur bacteria, along with Synechocystis sp. Sll0086, contain a highly conserved switch I and A-loop, whereas AtGet3d, NosGet3d, and Synechocystis sp. Slr1794 do not. It will be interesting to investigate the differences between the two Synechocystis sp. Get3d proteins as one is more similar to AtGet3d and NosGet3d than its paralog, which has features closer to cytoplasmic Get3 proteins implying distinct roles.
Catalytic residues show some interesting variability in Get3d. The canonical Walker A lysine, which coordinates the nucleotide \(\beta\)-phosphates, is broadly conserved in P-loop NTPases (Leipe et al., 2002). For Get3d, this lysine is conserved throughout angiosperms, such as A. thaliana. Green sulfur (Chlorobiaceae) and green nonsulfur (Chloroflexi) bacteria also have a lysine at this position (Figure 5.1B). Certain cyanobacterial Get3d proteins have replaced this with an arginine, which is unique to Get3d proteins that appear early in the cyanobacterial lineage, including simple filamentous cyanobacteria, such as some Pseudanabaena species, Leptolyngbya, Halomicronema, and several clades of synechococcalean cyanobacteria (Mareš et al., 2019; Moore et al., 2019). Based on this evidence, the Walker A lysine may have mutated early in the cyanobacterial lineage, whereas green sulfur and nonsulfur bacteria and angiosperms retained the lysine. The catalytic aspartate in Get3, D57 in yeast, coordinates water and primes it for nucleophilic attack of the \(\gamma\)-phosphate of ATP (Mateja et al., 2009; Suloway et al., 2009). This residue is highly conserved in Get3d proteins; however, it is a glutamate in NosGet3d, which may contribute to its lower Vmax (Figure 5.1B).
Evidence supports that A. thaliana has two Get3 paralogs localized to the chloroplast: Get3b and Get3d (Anderson et al., 2021; Xing et al., 2017). While AtGet3d appeared early in the evolution of photosynthesis, AtGet3b is first found with the appearance of chloroplasts, suggesting it was a newly acquired role in plants. A possible role is that AtGet3b and AtGet3d are both involved in TA protein targeting with different substrate specificities or different destination membranes (thylakoid versus inner envelope membrane). This is an exciting hypothesis, as AtGet3b was shown to interact with the thylakoid membrane protein AtSECE1 but not the inner envelope membrane AtSECE2 (Anderson et al., 2021). While previous work has shown that AtGet3b localizes to the chloroplast stroma specifically (Anderson et al., 2021), we were unable to distinguish between the stroma and thylakoid lumen here. Thus, the possibility that AtGet3b functions in the stroma and AtGet3d functions in the thylakoid lumen cannot be ruled out. AtGet3b and AtGet3d may also act in different tissues, in different plastid types (e.g., chloroplast, leucoplast, and chromoplast) (Choi et al., 2021), or during different stages of development. As they are both conserved across plants, it will be necessary to determine the roles of AtGet3b and AtGet3d.
If Get3d plays a role in TA protein targeting in chloroplasts, possible clients include several essential chloroplast-encoded proteins such as multiple photosystem I and II reaction center components and several cytochrome b6f proteins (Dobrogojski et al., 2020). The pool of substrates may also include nuclear-encoded TA proteins such as SECE1 and SECE2. Many of these proteins are conserved across all photosynthesis and represent an interesting pool of possible Get3d substrates.
The deep evolutionary connection between the Get3d fold and photosynthesis, while correlative, does not address the function of Get3d. Clearly, the preservation of its presumed function during the endosymbiotic event that created chloroplasts provides evidence that the role is critical for the conversion of light into chemical energy. This function remains to be determined, but as is seen for cytoplasmic Get3 proteins, it is likely either involved in TA protein targeting, acts as a chaperone, or perhaps both? A completely new role is possible, and future work to identify phenotypes and interaction partners should illuminate this puzzle. The breadth of Get3-like proteins scattered across kingdoms leads to questions about where this fold first appeared and what that role may have been.
5.5 Material and Methods
5.5.1 A reference alignment and phylogeny of the Get3/ArsA family
All proteins from UniProt version 2020_06 with an annotation as Get3/ArsA were pulled down (IPR027542, IPR016300, and IPR025723) along with all other InterPro domains identified (Blum et al., 2021; Mistry et al., 2021; UniProt Consortium, 2021). Get3/ArsA domains were then identified by searching against hidden Markov models for monomeric Get3/ArsA domains from solved crystal structures, split by hand for pseudodimers. The resulting domains were then searched (jackhammer, three iterations (Eddy and Wheeler, 2020)) against the preliminary set of UniProt proteins to find additional monomeric representatives. Each hit that covered 90% of the best scoring query was considered complete. Pseudodimers were split midway between the end of the first domain and the start of the second domain on the parent sequence.
A reference alignment of Get3/ArsA domains was then created by clustering domains using mmseqs at 65% sequence identity (Steinegger and Söding, 2018). Clusters were then aligned using mafft, version 7.471 (Katoh and Standley, 2013) with a seed alignment given by a structural alignment (STAMP) of Get3/ArsA proteins with solved structures (genafpair, maxiterate 1000, and retree 20) (Russell and Barton, 1992). A maximum-likelihood phylogeny was computed using RAxML, version 8.2.12 (Stamatakis, 2014): automatic model assignment using machine learning criterion, best scoring model LG with empirical base frequencies; and rapid bootstrap search complete after 400 replicates (-I autoMRE) (Pattengale et al., 2010).
5.5.2 Plant, photosynthetic, and \(\alpha\)CD containing Get3/ArsA proteins
Get3/ArsA proteins with \(\alpha\)CD domains were identified by a match to Pfam PF17886 (ArsA_HSP20) as annotated by UniProt. Organisms were defined as putatively photosynthetic if a UniProt proteome contained more than 10 proteins assigned to the Gene Ontology term GO:0015979 (photosynthesis) (Ashburner et al., 2000; Gene Ontology Consortium, 2021). Because of uneven sequencing coverage across genomes, analysis of the presence of \(\alpha\)CD-containing Get3 proteins in photosynthetic versus nonphotosynthetic organisms was carried out for proteomes annotated by UniProt as reference, nonredundant, or with a BUSCO completeness >75% (Simão et al., 2015).
Get3 proteins were separated into subsets to be clustered (mmseqs easycluster) into representative sequences for phylogenetic analysis with specific minimum sequence identity levels for each group: Get3 proteins from plants (85%), Get3 proteins with \(\alpha\)CD domains from photosynthetic bacteria (70%), Get3 proteins without \(\alpha\)CD domains from photosynthetic bacteria (65%), and all other Get3/ArsA proteins (60%). Where one of a pair of a pseudodimer was indicated as a representative, the other half was kept as well. Get3 proteins from A. thaliana and those with solved crystal structures were also included. The resulting sequences were then aligned using mafft (genafpair, maxiterate 1000, and retree 20) seeded by the structure-based alignment specified previously.
A maximum-likelihood phylogeny was computed using RAxML: automatic model assignment using ML criterion, best scoring model VT with fixed base frequencies; rapid bootstrap search complete after 100 replicates (-I autoMRE); N-terminal only, Get3-only, and C-terminal portion only tested as subpartitions. Nonrepresentative sequences were then added into the alignment using mafft (--add, --keeplength) and then placed onto the tree using the RAxML Evolutionary Placement Analysis (-f v) (Berger et al., 2011).
The resulting tree is post facto rooted using the most recent common ancestor of E. coli ArsA and MjGet3. Plant proteins are assigned to Get3a, b/c, or d by the presence of the corresponding A. thaliana protein in that clade. Trees are manipulated and drawn using phyloseq (McMurdie and Holmes, 2013), phytools (Revell, 2012), treeio (Wang et al., 2020), ggtree (Yu, 2022, 2020), tidytree (Yu, 2022), and ggplot2 (Wickham, 2016) packages in R (R Core Team, 2015) and the tidyverse (Wickham et al., 2019).
5.5.3 Cloning
The AtGet3d purification and crystallization construct was prepared by inserting AtGet3d \(\Delta\) 1–57 into the NdeI–XhoI cut sites of pET22b(+) using standard restriction enzyme cloning methods. Based on signal/chloroplast targeting peptide prediction (Almagro Armenteros et al., 2019) and mafft sequence alignments (Katoh and Standley, 2013), the first 57 amino acids were truncated from AtGet3d.
Get3d-GFP fusion constructs were generated by isothermal assembly following standard procedures (Gibson et al., 2009). Each assembled construct was in the pENTR plasmid with L1 and L2 sites that allowed for recombination of the Get3d-GFP construct into a destination vector with the UBQ10 promoter regulatory sequence and an OCS terminator sequence (pMOA pUBQ10-GW-OCS).
The AtGet3d ATPase assay construct was prepared using standard Gibson cloning protocols (Gibson et al., 2009) by inserting \(\textit{At}\text{Get3d}_{\Delta1-57}\) into a pET22b(+) vector containing an N-terminal His6 tag and human rhinovirus 3C protease cut site. The pET33b-His6-TEV-NosGet3d ATPase assay construct was prepared using standard Gibson cloning methods (Gibson et al., 2009), and pET33b-His6-TEV-ScGet3 was prepared as described previously (Suloway et al., 2009).
The Get3d TA protein capture assay constructs were prepared using standard Gibson cloning methods (Gibson et al., 2009). The MBP-Sbh1 TA protein was prepared as described previously (Lin et al., 2021).
5.5.4 Plant material
N. benthamiana seeds were germinated on Sunshine Mix 5 with perlite and vermiculite added at a ratio of 3:1:1, respectively. After seedlings germinated and the first true leaves appeared, plants were transplanted and allowed to grow for 14 days in 16:8 light:dark hour cycle.
Agrobacteria transformation and tobacco infiltration of Get3d-GFP variants
For transient expression, plasmids pUBQ10::Get3d-GFP and pUBQ10::Get3d\(\Delta\)TP-GFP were introduced into the Agrobacterium strain GV3101 by triparental mating. Agrobacteria strains were grown in 2xYT media with gentamycin (30 \(\mu\)g/ml), rifampicin (50 \(\mu\)g/ml), and spectinomycin (100 \(\mu\)g/ml) and adjusted to an absorbance of 0.1 in 10 mM MgCl2 and 150 \(\mu\)M acetosyringone. About 1 ml of each Agrobacterium sample was infiltrated into each leaf using a 1 ml syringe. Three to four leaves were infiltrated per construct.
5.5.5 Confocal microscopy
After 48 h, tobacco leaf samples were imaged on an upright Zeiss 780 Confocal Laser Scanning Microscope. The 488 nm laser line was used to excite both GFP and induce chlorophyll autofluorescence. Standard excitation and emission windows were used for GFP and chlorophyll b. Microscopy images were processed in ImageJ (Schindelin et al., 2012; Schneider et al., 2012).
5.6 Supplementary Figures